-
抗体試薬
- フローサイトメトリー用試薬
-
ウェスタンブロッティング抗体試薬
- イムノアッセイ試薬
-
シングルセル試薬
- BD® AbSeq Assay | シングルセル試薬
- BD Rhapsody™ Accessory Kits | シングルセル試薬
- BD® Single-Cell Multiplexing Kit | シングルセル試薬
- BD Rhapsody™ Targeted mRNA Kits | シングルセル試薬
- BD Rhapsody™ Whole Transcriptome Analysis (WTA) Amplification Kit | シングルセル試薬
- BD Rhapsody™ TCR/BCR Profiling Assays (VDJ Assays) | シングルセル試薬
- BD® OMICS-Guard Sample Preservation Buffer
- BD Rhapsody™ ATAC-Seq Assays
-
細胞機能評価のための試薬
-
顕微鏡・イメージング用試薬
-
細胞調製・分離試薬
-
- BD® AbSeq Assay | シングルセル試薬
- BD Rhapsody™ Accessory Kits | シングルセル試薬
- BD® Single-Cell Multiplexing Kit | シングルセル試薬
- BD Rhapsody™ Targeted mRNA Kits | シングルセル試薬
- BD Rhapsody™ Whole Transcriptome Analysis (WTA) Amplification Kit | シングルセル試薬
- BD Rhapsody™ TCR/BCR Profiling Assays (VDJ Assays) | シングルセル試薬
- BD® OMICS-Guard Sample Preservation Buffer
- BD Rhapsody™ ATAC-Seq Assays
- Japan (Japanese)
-
Change country/language
Old Browser
Looks like you're visiting us from {countryName}.
Would you like to stay on the current country site or be switched to your country?
.png)
.png)
Regulatory Statusの凡例
Any use of products other than the permitted use without the express written authorization of Becton, Dickinson and Company is strictly prohibited.
Preparation and Storage
推奨アッセイ手順
BD™ CompBeads can be used as surrogates to assess fluorescence spillover (Compensation). When fluorochrome conjugated antibodies are bound to CompBeads, they have spectral properties very similar to cells. However, for some fluorochromes there can be small differences in spectral emissions compared to cells, resulting in spillover values that differ when compared to biological controls. It is strongly recommended that when using a reagent for the first time, users compare the spillover on cell and CompBead to ensure that BD Comp beads are appropriate for your specific cellular application.
Product Notices
- This reagent has been pre-diluted for use at the recommended Volume per Test. We typically use 1 × 10^6 cells in a 100-µl experimental sample (a test).
- An isotype control should be used at the same concentration as the antibody of interest.
- Source of all serum proteins is from USDA inspected abattoirs located in the United States.
- Caution: Sodium azide yields highly toxic hydrazoic acid under acidic conditions. Dilute azide compounds in running water before discarding to avoid accumulation of potentially explosive deposits in plumbing.
- For fluorochrome spectra and suitable instrument settings, please refer to our Multicolor Flow Cytometry web page at www.bdbiosciences.com/colors.
- Please refer to http://regdocs.bd.com to access safety data sheets (SDS).
- Species cross-reactivity detected in product development may not have been confirmed on every format and/or application.
- Please refer to www.bdbiosciences.com/us/s/resources for technical protocols.
関連製品



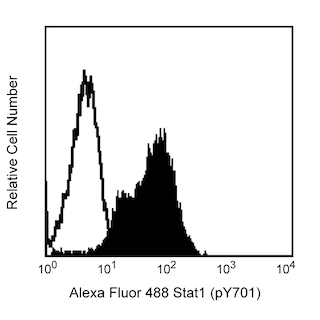

STATs (signal transducers and activators of transcription) are critical mediators of the biologic activity of cytokines including Interleukins (IL) 2-5, IL-7, IL-15, GM-CSF, erythropoietin and growth hormone. Ligand-receptor interaction leads to activation of constitutively associated JAK family kinases and subsequent recruitment/activation of STATs by tyrosine phosphorylation. Active STATs then move to the nucleus to promote transcription of cytokine-inducible genes. Seven STAT proteins have been cloned, each of which is differentially expressed and/or activated in a cytokine-specific and cell type-specific manner. Stat6 plays an important role in signaling pathways that lead to the differentiation of T helper type 2 (Th2) cells from uncommitted CD4 T cell precursors. Moreover, IL-4, secreted by activated T lymphocytes, basophils, and mast cells, induces specific gene expression via the induction of tyrosine phosphorylation of Stat6 at tyrosine 641 (Y641). The SH3:SH2 domain of Stat6 associates with tyrosine-phosphorylated IL-4 receptor and the proximal Jak kinase phosphorylates Stat6 at Y641 on the C-terminal side of the SH2 domain. Stat6 is then released from the receptor, dimerizes, and is thought to contact the basal transcription machinery by binding to p300/CBP. While Stat6 is widely expressed in human tissues, it exhibits elevated expression in peripheral blood lymphocytes, colon, intestine, ovary, prostate, thymus, spleen, kidney, liver, lung, and placenta.
The 23/Stat6 monoclonal antibody recognizes Stat6, regardless of phosphorylation status.
Development References (9)
-
Bartoli M, Gu X, Tsai NT, et al. Vascular endothelial growth factor activates STAT proteins in aortic endothelial cells. J Biol Chem. 2000; 275(43):33189-33192. (Clone-specific: Immunofluorescence, Immunoprecipitation, Western blot). View Reference
-
Bromberg J, Darnell JE. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000; 19(21):2468-2473. (Biology). View Reference
-
Dent AL, Hu-Li J, Paul WE, Staudt LM. T helper type 2 inflammatory disease in the absence of interleukin 4 and transcription factor STAT6. Proc Natl Acad Sci U S A. 1998; 95(23):13823-13828. (Biology). View Reference
-
Heim MH. The Jak-STAT pathway: specific signal transduction from the cell membrane to the nucleus. Eur J Clin Invest. 1996; 26(1):1-12. (Clone-specific). View Reference
-
Hou J, Schindler U, Henzel WJ, Ho TC, Brasseur M, McKnight SL. An interleukin-4-induced transcription factor: IL-4 Stat. Science. 1994; 265(5179):1701-1706. (Biology). View Reference
-
Mikita T, Campbell D, Wu P, Williamson K, Schindler U. Requirements for interleukin-4-induced gene expression and functional characterization of Stat6. Mol Cell Biol. 1996; 16(10):5811-5820. (Biology). View Reference
-
Quelle FW, Shimoda K, Thierfelder W, et al.. Cloning of murine Stat6 and human Stat6, Stat proteins that are tyrosine phosphorylated in responses to IL-4 and IL-3 but are not required for mitogenesis. Mol Cell Biol. 1995; 15(6):3336-3343. (Biology). View Reference
-
Waite KJ, Floyd ZE, Arbour-Reily P, Stephens JM. Interferon-gamma-induced regulation of peroxisome proliferator-activated receptor gamma and STATs in adipocytes. J Biol Chem. 2001; 276(10):7062-7068. (Clone-specific: Western blot). View Reference
-
Xia Z, Salzler RR, Kunz DP, et al. A novel serine-dependent proteolytic activity is responsible for truncated signal transducer and activator of transcription proteins in acute myeloid leukemia blasts. Cancer Res. 2001; 61(4):1747-1753. (Clone-specific: Western blot). View Reference
Please refer to Support Documents for Quality Certificates
Global - Refer to manufacturer's instructions for use and related User Manuals and Technical data sheets before using this products as described
Comparisons, where applicable, are made against older BD Technology, manual methods or are general performance claims. Comparisons are not made against non-BD technologies, unless otherwise noted.
For Research Use Only. Not for use in diagnostic or therapeutic procedures.
Report a Site Issue
This form is intended to help us improve our website experience. For other support, please visit our Contact Us page.