-
Training
- Flow Cytometry Basic Training
-
Product-Based Training
- BD Accuri™ C6 Plus Cell Analyzer
- BD FACSAria™ Cell Sorter Cell Sorter
- BD FACSCanto™ Cell Analyzer
- BD FACSDiscover™ A8 Cell Analyzer
- BD FACSDiscover™ S8 Cell Sorter
- BD FACSDuet™ Sample Preparation System
- BD FACSLyric™ Cell Analyzer
- BD FACSMelody™ Cell Sorter
- BD FACSymphony™ Cell Analyzer
- BD LSRFortessa™ Cell Analyzer
- Advanced Training
Old Browser
This page has been recently translated and is available in French now.
Looks like you're visiting us from {countryName}.
Would you like to stay on the current location site or be switched to your location?
Fragment-Seq: A Spatial Transcriptomics Method to Study Tissue Microenvironments at Single-Cell Resolution
A Novel Spatial Transcriptomics Method Based on Cellular Communities
Spatial transcriptomics is a rapidly evolving field aiming to capture transcriptomic features and spatial relationships of cells within tissues. Current methods have limitations such as the need for prior knowledge for targeted panel assembly, lack of single-cell resolution or low throughput.1
A novel method developed by researchers led by Andreas Moor at ETH Zürich and published in Nature Communications in November 2023 overcomes these limitations by enabling unsupervised, high-throughput and single-cell resolution analysis of spatially distinct tissue niches. This method utilizes existing single-cell sequencing platforms such as the BD Rhapsody™ Single-Cell Analysis System and is a powerful tool to investigate differences in cellular composition and gene expression between distinct tissue niches and serves as a valuable addition to currently available technologies in the field of spatial transcriptomics
Unveiling Local Tissue Microenvironment
In this study, Handler et al. developed fragment-seq, a novel method that combines partial tissue dissociation, size-dependent sorting of cellular communities (fragments) with a large fragment biosorter, cell hashing with lipid-tagged barcodes and single-cell RNA-sequencing. By exploiting the spatial proximity of landmark genes or cell types, fragment-seq allows the reconstruction of the native position of fragments within tissues, such as liver zonation or metastatic niches.
Fragment-seq was applied to dissect spatially distinct niches in a metastasis-bearing mouse liver, highlighting the importance of immune zonation and the tumor microenvironment. Using a previously identified liver endothelial cell zonation-specific gene expression signature, the researchers reconstructed the native position of a fragment along the central-portal axis. They identified differentially expressed genes in liver endothelial cells and Kupffer cells from different zones. The zonation patterns of metabolic genes and ligand-receptor interactions were validated using molecular cartography, a highly multiplexed fluorescence in-situ hybridization approach. Fragments were grouped based on their relation to metastatic sites, defined as ‘proximal’ if they contained metastatic cells and ‘distal’ if they did not. Proximal areas had a significantly higher proportion of macrophages/monocytes and metastatic cells, while the number of Kupffer cells and liver endothelial cells were reduced. An increased abundance of C1q-expressing macrophages was observed in metastatic-proximal sites, which are reportedly involved in T cell exhaustion and poor prognosis in many cancers. Many spatially enriched interactions involving macrophage/monocyte-derived ligands and integrin receptors on T cells were revealed, which might be involved in promoting tumorigenesis.
The researchers also demonstrated that fragment-seq can be easily adapted to other tissue types and species with minor adjustments to the single-cell dissociation protocol. They created proof-of-principle datasets from the mouse spleen and human Crohn’s disease biopsies. For the latter, they introduced size selection based on sequential filtering and manual picking of fragments to make fragment-seq feasible also in the absence of an expensive biosorter apparatus. They showed that fragment-seq can distinguish fibrotic and non-fibrotic microenvironments in Crohn’s disease samples based on the presence or absence of activated fibroblasts.
Fragment-seq is a powerful tool to investigate differences in cellular composition and gene expression between distinct tissue niches and serves as a valuable addition to currently available technologies in the field of spatial transcriptomics. It combines the advantages of single-cell resolution, high throughput and unsupervised spatial hypothesis generation for entire tissues.
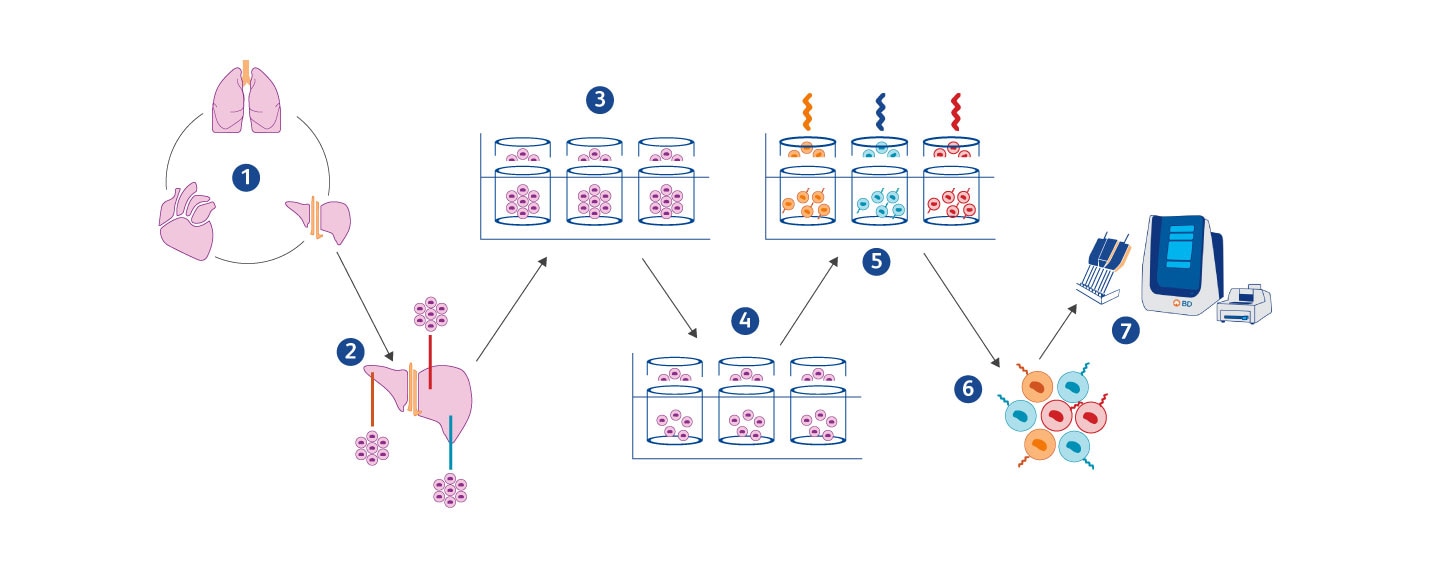
(1) After PBS perfusion, multiple organs are collected from euthanized mice. (2) Tissues are partially dissociated into fragments using the mechanical force of a scissor. (3) Individual fragments of Ø 200–450 µm are sorted with a large fragment biosorter into a 96-well plate. (4) Subsequently, fragments are dissociated into single cells within each well and (5) hashed with a fragment-specific lipid-tagging barcode to preserve information about a cell’s neighborhood prior to (6) pooling for scRNA-seq. (7) Whole transcriptome analysis (WTA) on MULTI-seq labeled cells is performed using the BD Rhapsody™ Single-Cell Analysis System.
Read the full publication entitled, “Fragment-sequencing unveils local tissue microenvironments at single-cell resolution.”
References
1. Dario Bressan et al. The dawn of spatial omics.Science381,eabq4964(2023).DOI:10.1126/science.abq4964
For Research Use Only. Not for use in diagnostic or therapeutic procedures.