-
Training
- Flow Cytometry Basic Training
-
Product-Based Training
- BD Accuri™ C6 Plus Cell Analyzer
- BD FACSAria™ Cell Sorter Cell Sorter
- BD FACSCanto™ Cell Analyzer
- BD FACSDiscover™ A8 Cell Analyzer
- BD FACSDiscover™ S8 Cell Sorter
- BD FACSDuet™ Sample Preparation System
- BD FACSLyric™ Cell Analyzer
- BD FACSMelody™ Cell Sorter
- BD FACSymphony™ Cell Analyzer
- BD LSRFortessa™ Cell Analyzer
- Advanced Training
Old Browser
This page has been recently translated and is available in French now.
Looks like you're visiting us from {countryName}.
Would you like to stay on the current location site or be switched to your location?
A Novel Assay to Evaluate Single-Cell Sorting Accuracy Using Antibody-Derived Tag-Based qPCR
Development of single-cell technologies has played a vital role in unraveling cell type heterogeneity, revealing cellular functions and improving our understanding of complex biological systems over the last decade. Single-cell sorting is one of the key methods to facilitate downstream characterization of individual cells. It is a powerful tool to isolate individual cells into 96- or 384-well plates with information regarding the location and fluorescence intensity saved for each well. When single-cell sorting is integrated with downstream sequencing applications, users can link the protein expression phenotype to gene expression, which enables deep profiling and improved understanding of the function of the sorted single cells.
Even though single-cell sorting is reported to have high accuracy on different sorting instruments,1 operation of a cell sorter can be technically challenging. While sorting many plates simultaneously, users would need a simple and cost-effective assay to validate the sorting accuracy on selected sort events. Shi et al., provide a solution to evaluate single-cell sorting accuracy by leveraging antibody-derived tags (ADTs) and downstream TaqMan qPCR.
The cells were first co-stained with the same marker of both ADTs and fluorescent antibodies. After single-cell sorting, the oligo of the ADT reagent was amplified as a surrogate signal for the protein expression using multiplex TaqMan qPCR on sorted cells. This assay was then used not only to confirm the identity of the sorted events but also to characterize their proteomic signature at the single-cell level. Results showed that the assay was very sensitive, in that it could even differentiate doublets from single cells. Finally, this assay was modified to detect protein expression on lysed whole cells and the results demonstrated that the assay could detect protein levels from a small number of cells, an advantage over the existing protein detection methods such as western blotting.
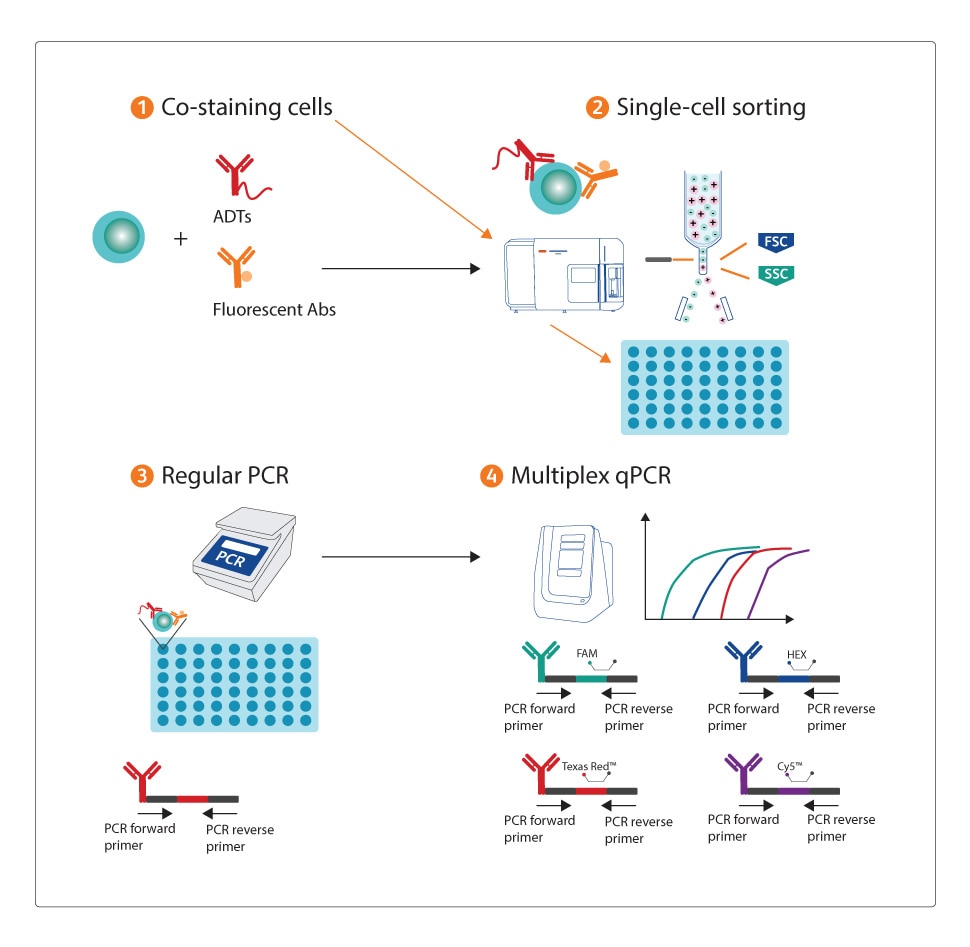
The study used the BD FACSDiscover™ S8 Cell Sorter for isolating single cells into a plate. The instrument provided information not only of the location and fluorescence intensity, but also images of every sorting event. Those images provided additional information to confirm the identity of sorting event. All the ADT reagents used were BD® AbSeq Antibody-Oligos, which are compatible with BD Rhapsody™ Single-Cell Analysis System. Flow cytometry data analysis was performed using either BD FACSChorus™ or FlowJo™ (V10.10.0) Software.
This novel ADT-based PCR assay reported by Shi et al. can be used to validate the accuracy of cell sorting along with detection of protein expression at a single-cell level.
Read the full paper entitled, “Evaluation of single-cell sorting accuracy using antibody-derived tag-based qPCR.”
References
- Penter L, Dietze K, Bullinger L, Westermann J, Rahn, H et al. FACS single cell index sorting is highly reliable and determines immune phenotypes of clonally expanded T cells. European J of Immunol. 2018; https://doi.org/10.1002/eji.201847507
For Research Use Only. Not for use in diagnostic or therapeutic procedures