-
Training
- Flow Cytometry Basic Training
-
Product-Based Training
- BD Accuri™ C6 Plus Cell Analyzer
- BD FACSAria™ Cell Sorter Cell Sorter
- BD FACSCanto™ Cell Analyzer
- BD FACSDiscover™ A8 Cell Analyzer
- BD FACSDiscover™ S8 Cell Sorter
- BD FACSDuet™ Sample Preparation System
- BD FACSLyric™ Cell Analyzer
- BD FACSMelody™ Cell Sorter
- BD FACSymphony™ Cell Analyzer
- BD LSRFortessa™ Cell Analyzer
- Advanced Training
Old Browser
This page has been recently translated and is available in French now.
Looks like you're visiting us from {countryName}.
Would you like to stay on the current location site or be switched to your location?
The BD Cellismo™ Data Visualization Tool is a software tool for secondary analysis and visualization of single-cell multiomics data. It helps researchers turn millions of data points into actionable insights without writing a single line of code.
With the BD Cellismo™ Data Visualization Tool, you can easily visualize data from your samples across various parameters; subset cells into populations based on gene expression; create histograms, heatmaps, dot plots, volcano plots, violin plots, tSNE and UMAP plots; and customize and export plots as .PNG or .SVG files for collaboration and publication.
Watch the video to see how you can analyze your data using both primary analysis, with the BD Rhapsody™ Sequence Analysis Pipeline, and secondary analysis.
Features
-
Intuitive: Point and click to visualize your data as graphs and plots
-
User Friendly: No cost of adoption; no coding required
-
Multiomics Enabled: Explore single-cell RNA, protein and ATAC datasets with ease
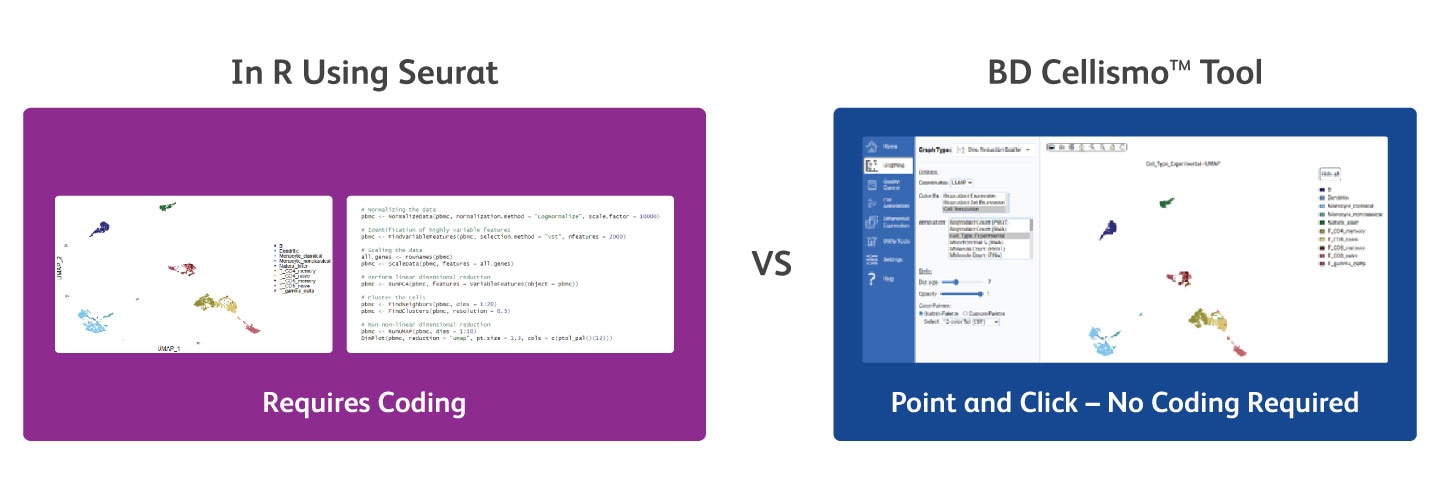
Generate the Same Data Visualization as Seurat Using the BD Cellismo™ Data Visualization Tool—Without Writing Any Code or Programming
With the BD Cellismo™ Data Visualization Tool (right) you’re able to generate the same data visualization as with Seurat (left), without having to write any code. The BD Cellismo™ Data Visualization Tool’s interface also lets you easily customize your figures (e.g., dot size, color, opacity) in a point and click fashion.
Input Requirements:
For WTA or AbSeq data:
- (Preferred) .Cellismo output file from the BD Rhapsody™ Sequence Analysis Pipeline.
- Alternative input formats:
- Market exchange (MEX)
- H5MU/H5AD
- Seurat .rds
- Market exchange (MEX) compressed with .zip or tar.gz
- H5
- Cellismo output file from the BD Rhapsody™ Sequence Analysis Pipeline (V3 or later only)

Release Notes
Latest Version: v2.0 | Release Date: October 2025
Added:
- Support for BD Rhapsody™ ATAC-Seq Assay
- New modalities: ATAC_PEAKS, ATAC_GENE_ACTIVITY, ATAC_MOTIF
- Genome browser to view ATAC peaks and ATAC fragments in the context of genomic locations and genes
- Motif enrichment analysis tool—find transcription factor motifs that are enriched in a given set of peaks, compared to background peaks
- Peak and RNA expression correlation tool—find peaks whose activity correlate with RNA expression
- RNA vs ATAC heatmap—see RNA expression and ATAC gene activity correlation for a given cell annotation
- Ability to combine and reanalyze multiple ATAC datasets
- Doublet detection tool
- Batch correction utility tool, with batch correct counts available to be used in graphing pages
- Integration utility tool
- New cell classification tool, ScType, with the ability to customize marker genes
- Confidence score annotations for cell type classification tools
- Project action recording on the current project page
- Data export options—for R / Seurat, Scanpy (H5AD), Muon (H5MU), FCS and CSV
- RNA data import option—Seurat RDS
- New DESeq2 method in the Differential Analysis tool
- "Selected Cell Actions" menu on graphing pages, and added "Delete Cells" and "Quick Differential Analysis" options
- Switch to reverse color palette
- Contour lines overlay to biaxial plot
- Options to start project, including flavor of highly variable gene selection, number of highly variable genes and number of PCA/LSI components
- Ability to filter bioproduct dropdown by modality
- Ability to Export and restore app settings
Updated:
- Renamed Differential Expression tool to Differential Analysis and added support for all ATAC modalities
- Cell annotation—Clustering tool to support ATAC modalities
- Dimensionality Reduction utility tool to support ATAC modalities
Fixed:
- Error in t-SNE generation leading to poor dimensionality reduction results
- Overall QC failure when no mitochondria or ribosomal genes are present
- Start project failure when only 1 prot marker is present
System requirements
- Windows 10 or later (64-bit)
- MacOS 14 (Sonoma) or later (Intel or Apple silicon chip)
- 8 GB RAM minimum
RNA/AbSeq
- 16 GB RAM for 200,000 cells
- 64 GB RAM for 1M cells
ATAC alone or ATAC multiomic
- 16 GB RAM for 100,000 cells
- 64 GB RAM for 500,000 cells
Free, but Powerful–Try it Out for Yourself
Download the free BD Cellismo™ Data Visualization Tool today. After you provide your information and click Submit, download will begin automatically. We will use your information to keep you updated on latest developments, updates and bug fixes as they become available.
Windows Users:
Once the file has downloaded, double-click the .exe file. You will see a simple animation as it installs. After the install is complete, the program will open automatically. A shortcut will be placed on your desktop.
Mac Users:
There are two different install packages depending on your Chip architecture—Mac (Intel) or Mac (Apple M). To check your chip architecture, press the Apple menu on the top left. Select About This Mac; where it says Chip, it should say either Intel or Apple. In the form below, pick the option that applies to your system. Once the file has downloaded, double-click the .zip file to extract Cellismo.app. Double-click Cellismo.app to open it. The app can be moved to the folder of your choice (like the user's Applications directory).
Once installed, start new projects on the Home page under the Start New Project tab. Drag-and-drop data files onto the page and click Start Analysis.
Get a step-by-step demonstration of the new capabilities of the BD Cellismo™ Tool, including support for BD Rhapsody™ ATAC-Seq Assays, batch correction, doublet detection and cell classification using the new ScType algorithm.
Get a step-by-step demonstration of the capabilities of the BD Cellismo™ Tool, a new software tool designed to help you easily visualize multiomic single-cell data.
Learn how to import single-cell data into this tool. Drag and drop .cellismo, .h5mu, .h5ad or mex formats. Explore demo data from PBMC with 2,500 cells, 30 proteins and 2,500 genes.
Learn how to use the BD Cellismo™ Tool’s lasso selection tool to select cells based on protein expression. Explore biaxial graphs showing CD3, CD4 and CD45RA expression. Select CD4 T cells, create new annotations and identify CD4 naïve and memory T cells.
Discover the seven graph types available in the tool for data exploration. Use t-SNE and UMAP for dimensionality reduction. Customize graphs with adjustable dot size, opacity and color palettes. Save plots as SVG or PNG or add to the gallery. Export all graphs as PNG at the end of your analysis.
Download the Free BD Cellismo™ Tool
After you provide your information and click the Submit button, download will begin automatically.
*Required fields
For Research Use Only. Not for use in diagnostic or therapeutic procedures.
Apple and Mac are trademarks of Apple, Inc. Illumina is a trademark of Illumina, Inc. Intel is a trademark of Intel Corporation or its subsidiaries.








