-
Reagents
- Flow Cytometry Reagents
-
Western Blotting and Molecular Reagents
- Immunoassay Reagents
-
Single-Cell Multiomics Reagents
- BD® OMICS-Guard Sample Preservation Buffer
- BD® AbSeq Assay
- BD® OMICS-One Immune Profiler Protein Panel
- BD® Single-Cell Multiplexing Kit
- BD Rhapsody™ ATAC-Seq Assays
- BD Rhapsody™ Whole Transcriptome Analysis (WTA) Amplification Kit
- BD Rhapsody™ TCR/BCR Next Multiomic Assays
- BD Rhapsody™ Targeted mRNA Kits
- BD Rhapsody™ Accessory Kits
- BD® OMICS-One Protein Panels
-
Functional Assays
-
Microscopy and Imaging Reagents
-
Cell Preparation and Separation Reagents
-
Thought Leadership
- News
- Blogs
-
Scientific Publications
-
Events
- Expanding PARADIGM to Infectious Disease Modeling: HIV & Tuberculosis
- CYTO 2023: Advancing the World of Cytometry
- Advances in Immune Monitoring Series
- Validating Flow Cytometry Assays for Cell Therapy
- Enhancing Cell Analysis with a New Set of Eyes
- BD Biosciences at International Clinical Cytometry Society 2025
Old Browser
This page has been recently translated and is available in French now.
Looks like you're visiting us from {countryName}.
Would you like to stay on the current location site or be switched to your location?
A Novel Multiplex Flow Proxy Assay for the Simultaneous Measurement of Multiple Biomarkers
Combining the analysis of protein biomarkers to genome-wide expression profiling at the single-cell level provides deeper scientific insights than previously possible. Despite being widely adopted, CITE-Seq (Cellular Indexing of Transcriptomes and Epitopes by sequencing), the technique that enables this combination, still needs standardization and simplification to help overcome some of its challenges such as the lack of quality checks to test the binding of antibodies to report expression of biomarkers before proceeding with the whole CITE-Seq assay.
Usually, single-cell multiomic protocols involve specifically isolating the cell population of interest with fluorescence activated cell sorting (FACS) and then using that population for simultaneous single-cell sequencing of gene (RNA-Seq) and protein expression (CITE-Seq). This requires co-staining of cells with antibodies conjugated to both fluorescent dyes for flow cytometry analysis and sorting and oligonucleotides for CITE-Seq. This means that, in addition to the already mentioned uncertain binding of oligo-conjugated antibodies, this procedure also poses the risk of antibody competition and eventual compromise of the reporting signal intensity.
Shi et al. addressed these issues in a recent publication and provided an easy-to-use workflow utilizing a select set of fluorescent dye-oligo conjugates complementary to the specific antibody-derived tags (ADTs) used to perform the CITE-Seq assay. The strategy was called a flow proxy assay since it allowed confirmation of ADT binding to cells using flow cytometry prior to the single-cell capture, library preparation and sequencing workflows. As a proof of principle, the authors applied this method to different experimental scenarios from a single ADT marker detection up to a 10-marker multiplex detection approach, evaluating multiple biomarkers simultaneously. The authors also demonstrated the applicability of this strategy to selectively sort natural killer (NK) and T cells using a 5-marker panel of dye-oligo conjugates and fluorescent activated cell sorting followed by CITE-Seq.
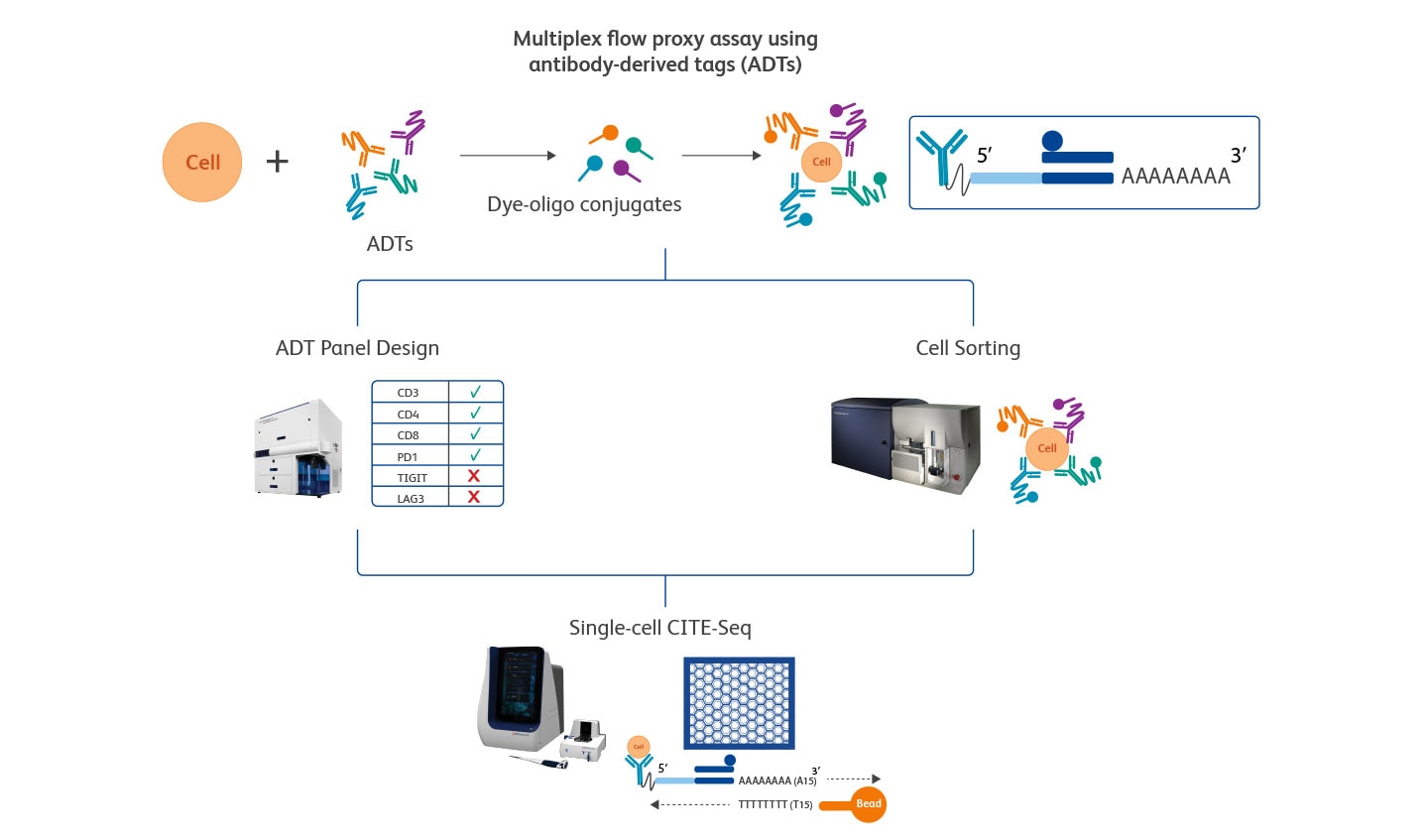
Among the technologies and materials that were used, BD FACSymphony™ A5 or BD FACSymphony™ A5 SE Cell Analyzers were applied for conventional or spectral flow cytometric analysis, respectively. A BD FACSAria™ II Cell Sorter was used for FACS. Multiple antibodies and dyes were needed including ready-to-use commercialized formats, customized conjugates and some of the newer BD Horizon RealYellow™ and BD Horizon RealBlue™ Dyes. FlowJo™ Software v10.8.1 was used for the visualization and analysis of flow cytometry data. The microwell-based BD Rhapsody™ Single-Cell Analysis System was used in combination with BD Rhapsody™ Targeted mRNA Kits, BD® AbSeq Antibody-Oligonucleotide Conjugates and BD® Single-Cell Multiplexing Kits for the multiomic single-cell analysis workflow.
In this publication, the authors introduce a flow cytometry–based assay to help users examine protein marker expression using antibody-derived tags (ADTs) for CITE-Seq prior to the single-cell capture, library preparation and sequencing workflows. Considering the different scenarios that were demonstrated, the flow proxy assay could be effectively used as a quality control while choosing the markers for ADT panels as well as a feasible option for experimental strategies that require fluorescence activated cell sorting followed by CITE-Seq.
Read the full paper entitled, “Flow cytometry analysis of protein expression using antibody-derived tags followed by CITE-Seq.”
BD Flow Cytometers are Class 1 Laser Products.
For Research Use Only. Not for use in diagnostic or therapeutic procedures.