-
Reagents
- Flow Cytometry Reagents
-
Western Blotting and Molecular Reagents
- Immunoassay Reagents
-
Single-Cell Multiomics Reagents
- BD® OMICS-Guard Sample Preservation Buffer
- BD® AbSeq Assay
- BD® Single-Cell Multiplexing Kit
- BD Rhapsody™ ATAC-Seq Assays
- BD Rhapsody™ Whole Transcriptome Analysis (WTA) Amplification Kit
- BD Rhapsody™ TCR/BCR Next Multiomic Assays
- BD Rhapsody™ Targeted mRNA Kits
- BD Rhapsody™ Accessory Kits
- BD® OMICS-One Protein Panels
- BD OMICS-One™ WTA Next Assay
-
Functional Assays
-
Microscopy and Imaging Reagents
-
Cell Preparation and Separation Reagents
Old Browser
This page has been recently translated and is available in French now.
Looks like you're visiting us from {countryName}.
Would you like to stay on the current location site or be switched to your location?
Single-Cell Multiomics
Single-cell multiomics (scM) is a cutting-edge approach that promises to revolutionize how we perceive and dissect cellular biology. Multiomics, the integration of genomic, transcriptomic, proteomic and other omics data, might initially seem redundant. Why go through the trouble of integrating multiple layers of molecular information when one omic could theoretically capture it all? The answer lies in the complexity of biological networks. Each omic readout provides valuable insights, but no single “ome” can fully unravel the intricate web of cellular processes. For example, while DNA dictates the potential of a cell, not all genes are transcribed into RNA and not all RNA is translated into proteins. Each criterion provides more information about the overall functioning of the cell.
Why Multiomics Matters Now
Multiomics methods offer more robust classifiers for biological samples, aiding in tasks such as cancer subtype identification and patient stratification. Beyond classification, multiomics provides a deeper understanding of genotype-phenotype relationships, unlocking insights into the fundamental mechanisms underlying biological traits and diseases.
In recognition of its significance, single-cell multimodal omics was named Nature Method of the Year in 2019, underscoring its pivotal role in advancing biological research. As we delve deeper into the realm of single-cell multiomics, we unlock new vistas of understanding, paving the way for breakthroughs in personalized medicine, disease treatment and beyond.
Check out BD tools for single cell-multiomics instruments, scM reagents and scM informatics analysis solutions.
The Five Omes
Genomics
Genomics is the study of an organism's DNA by deciphering the sequence of nucleotide bases comprising an organism's genome. Researchers gain invaluable insights into genetic variation, heredity and evolutionary relationships.

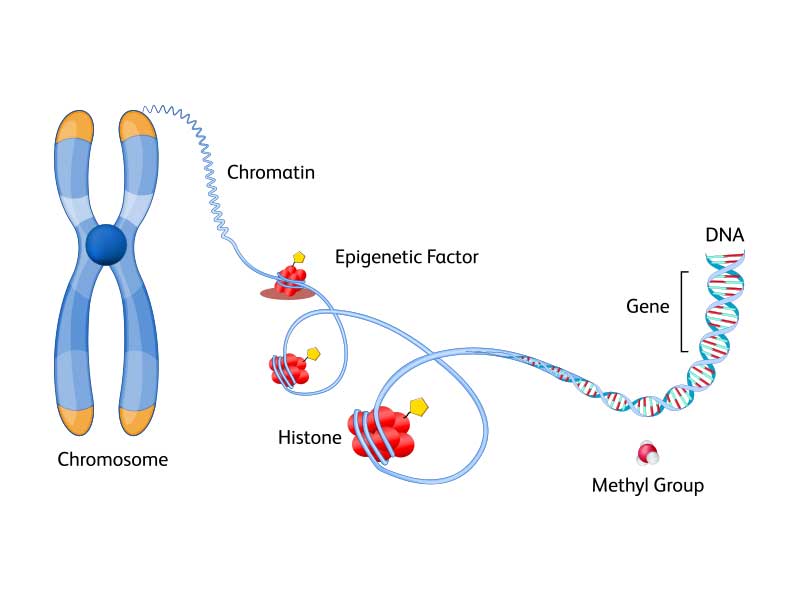
Epigenomics
Epigenomics explores the dynamic modifications to DNA and associated proteins that regulate gene expression without altering the underlying genetic code. Epigenetic mechanisms such as DNA methylation, histone modification and non-coding RNA regulation play pivotal roles in orchestrating cellular differentiation, development and response to environmental cues.
Explore BD solutions for single-cell epigenomics.
Transcriptomics
Transcriptomics delves into the vast landscape of RNA molecules within a cell or tissue, capturing the dynamic repertoire of gene expression patterns under various conditions.
Explore BD solutions for single-cell transcriptomics for the whole transcriptome or targeted mRNA.
Proteomics
Proteomics investigates the entire complement of proteins present within a biological sample. Proteins are the workhorses of the cell, executing diverse functions ranging from catalysis and signaling to structural support and defense. Proteomic analyses offer insights into protein abundance, post- translational modifications and protein-protein interactions.
Explore BD solutions for single-cell proteomics for cell surface proteins and intracellular proteins.
Metabolomics
Metabolomics is the study of small molecule metabolites that serve as the building blocks of cellular metabolism. Metabolites encompass a diverse array of compounds, including amino acids, lipids, sugars and neurotransmitters. Metabolomics elucidates metabolic pathways, metabolic fluxes and metabolic signatures associated with physiological states, environmental exposures and disease phenotypes.
Learn more about the latest trends in single cell multiomics on our blog page.

Discover how researchers are leveraging Single-Cell Multiomics in groundbreaking research to advance the field of biology
Immuno-Oncology
- Chronic lymphocytic leukemia (CLL)—This study demonstrates the power of single-cell multiomics in unraveling the complexities and heterogeneity of CLL, providing valuable insights for understanding its pathogenesis and potentially informing treatment strategies. Read the datasheet entitled, “Exploring tumor heterogeneity of chronic lymphocytic leukemia using single cell multiomics.”
- B cell lymphoma—This study delves into the interplay between malignant B cells and immune cells in B cell lymphoma's tumor microenvironment, offering insights with which to identify therapeutic targets in this disease. Read the datasheet entitled, “Leveraging the power of high-parameter cell sorting and single-cell multiomics to profile intratumoral immune cells in a model of B-cell lymphoma.”
- Melanoma—This study investigates tumor induced perturbations in cancer-related hematopoiesis, focusing on how tumors affect different hematopoietic stem and progenitor cell (HSPC) populations. Read the datasheet entitled, “Assessing tumor-driven perturbations during hematopoiesis in a melanoma mouse model through single-cell multiomic analysis.”
- Colorectal cancer— A study in Cell Reports Medicine entitled, “Human tumor-infiltrating MAIT cells display hallmarks of bacterial antigen recognition in colorectal cancer” found a specific subset of tumor infiltrating MAIT cells that respond to microbial antigens in colorectal cancer.1 Understanding these cells could pave the way to manipulate them or the microbiome for cancer therapy.
Download our application ebook, Advancing immuno-oncology through single-cell multiomics, to see how single-cell multiomics, the BD Rhapsody™ System and cell sorting can be used to gain important insights into cancer immunology.
Autoimmunity
- Lupus—A study entitled “Chronic immune activation in systemic lupus erythematosus and the autoimmune PTPN22 Trp620 risk allele drive the expansion of FOXP3+ regulatory T cells and PD-1 expression” found that SLE patients have more CD4+FOXP3+ cells due to thymically-derived Tregs expansion linked to PD-1, IL-2, and soluble SIGLEC-11.2
- Rheumatoid arthritis (RA)—A Nature Medicine paper entitled “Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis” delves into the cellular and molecular mechanisms of sustained remission in RA. It highlights distinct synovial tissue macrophage subsets that regulate inflammation and remission, revealing potential therapeutic targets to encourage resolution of inflammation and restore synovial homeostasis in RA.3
- Autoimmune demyelination—A study published in PNAS reveals that TNF-α signaling is crucial in peripheral nerve inflammation and autoimmunity. Aire-deficient mice exhibit increased TNF-α signaling, which recruits and activates immune cells, exacerbating autoimmune demyelination. Blocking TNF-α or its receptors ameliorates neuropathy, suggesting anti-TNF-α therapies could treat inflammatory neuropathies like GBS and CIDP.4
- Type 1 diabetes mellitus (T1DM)—A study investigated the T cell receptor (TCR) diversity in Japanese patients with T1DM using single-cell VDJ sequencing and showed distinct TCR sequences in autoreactive T cells and differences in gene expression of CD8+ and FOXP3+ cells compared to healthy subjects, suggesting altered TCR diversity and immune response in T1DM.5

Infectious Diseases
- Acute otitis media—In a rat model of acute otitis media, single-cell transcriptomic profiling revealed dual-feature cells related to phagocytosis. These cells coexisted with professional phagocytes and influenced inflammation progression. Macrophages played a central role in the inflammatory response, with M2 polarization affecting immune microenvironments.6
- COVID-19—A study identifies HIF1A as a long-term immunological scar in monocytes of COVID-19 convalescents without comorbidities. It correlates with disease severity, suggesting its potential as a biomarker for immune monitoring and treatment strategy design post-COVID-19, especially in symptom-free recovery phases.7
- Sepsis—Another study investigates mitochondrial STAT3’s role in macrophages during sepsis. It reveals that mitochondrial STAT3 exacerbates sepsis by promoting fatty acid oxidation (FAO) through CPT1a stabilization, mediated by USP501.8
- Tuberculosis-In this study, γδ T cells from a latent tuberculosis donor were enriched and analyzed using single cell immune profiling. It highlights the identification of full-length γδ T cell receptor sequences and clonal expansion, crucial for understanding the immune response to tuberculosis. Read this technote entitled "Single-cell T cell receptor repertoire analysis of sorted γδ T cells using the BD Rhapsody™ TCR/BCR Multiomic Assay".
Immune Cell Biology
- Circulating innate lymphoid cells (ILCs)—This study highlights the heterogeneity of ILCs, identified through single-cell multiomic analysis, and the development of high-parameter flow cytometry panels to study ILC subsets across different donors, revealing unique immunophenotypic signatures. Read the datasheet entitled “Resolving human circulating innate lymphoid cell heterogeneity using advanced single cell multiomic analysis.”
- Regulatory T cells (Tregs)—This study details the use protein and mRNA single-cell analysis, revealing distinct Treg differentiation states. It showcases the precise resolution of Treg subsets, advancing our understanding of their role in immune homeostasis and disease. Read the datasheet entitled “A comprehensive characterization of regulatory T cells using BD Rhapsody™ Single-Cell Analysis System.”
- Neutrophils—This study highlights that neutrophils and emergency granulopoiesis are central to the immune suppression observed in sepsis. Single-cell multiomics revealed distinct populations of immunosuppressive neutrophils and altered granulopoiesis in sepsis patients, suggesting potential targets for therapy and personalized medicine approaches in severe infections.9
- CD8+ T cells—This study describes a novel method called TetTCR-SeqHD, which profiles antigen-specific CD8+ T cells by integrating TCR sequencing with antigen specificity, gene expression and phenotyping. This high-throughput approach offers precise detection of antigen specificity and cross-reactivity, aiding in the study of T cell responses in diseases like type 1 diabetes.10
Immunotherapy
- Human head and neck squamous cell carcinomas (HNSCC)—In this study investigating immune alterations in HNSCC and inflamed tissues, researchers found that tumor-unique immune changes exist alongside general inflammatory patterns. Notably, intratumoral IL1R1+ regulatory T (T reg) cells exhibited superior suppressive function compared to IL1R1- T reg cells. Tumor-unique immune alterations, including the role of T reg cells, could have implications for immunotherapy strategies.11
- Lung adenocarcinoma—This study discusses the tumor immune microenvironment (TME) in EGFR mutant lung adenocarcinoma specifically the effect of lower PD-L1 expression and tumor mutation burden (TMB). The study uses single-cell transcriptome analysis to reveal a suppressive TME, with a lack of CD8+ tissue-resident memory cells and insufficient crosstalk between T cells and other cell types.12
- Adoptive T cell therapy (ACT)—This study discusses enhancing ACT for cancer treatment. The researchers found that combining ACT with transient anti-CD4 treatment increases the presence of IL-18Rαhi CD8+ T cells, which improves tumor suppression. This approach could potentially advance immunotherapy research.13
- Pancreatic ductal adenocarcinoma (PDAC)—This study discusses the role of innate immunity in PDAC, focusing on neutrophils within the tumor microenvironment. It highlights a study revealing various neutrophil subpopulations, including a pro-tumor type associated with poor prognosis. The findings suggest targeting these neutrophils could enhance immunotherapy for PDAC, offering an alternative to T cell-based treatments.14
Neuroscience
- Fetal brain development—This study focuses on microglia's role in fetal brain development. It reveals that embryonic microglia accumulate at critical cortical boundaries, preventing cavitary lesions caused by morphogenetic stress. The study highlights microglia's protective functions and the importance of the ATM-factor Spp1 in maintaining structural integrity during brain morphogenesis.15
- Neuropeptide Y (NPY)—The article discusses the role of NPY in regulating immune responses. It highlights how NPY, produced by neurons in the suprarenal and celiac ganglia, can modulate splenic immune activity. The study reveals that NPY influences lymphocyte proliferation and activation, suggesting its potential in treating inflammatory and autoimmune diseases. This neuron-immune system interaction is an ancient mechanism for maintaining immune balance.16
- FGF21 interactions with glutamatergic neurons—This study discusses the hormone FGF21, produced by the liver, and its role in suppressing sugar intake and preference by acting on glutamatergic neurons in the hypothalamus. The study reveals FGF21's direct action in the ventromedial hypothalamus (VMH) affects neuronal activity, specifically regulating sucrose intake without impacting non-nutritive sweet-taste preference, body weight or energy expenditure.17
- Alzheimer's disease (AD)—The article discusses the impact of the ABI3 gene on AD pathology in a mouse model. It reveals that ABI3 deletion increases amyloid β (Aβ) accumulation and impairs microglial function, leading to exacerbated neuroinflammation and synaptic dysfunction. This suggests ABI3's role in AD progression and potential as a therapeutic target.18
Plant Sciences
- Floral transition - The study identifies three stages of floral transition, highlights the erratic flowering in woody fruit crops like litchi due to environmental factors, and uses snRNA-seq to reveal gene expression patterns crucial for understanding bud development and flowering 19
- Organ regeneration - The study reveals that WOX5/7 proteins enhance auxin production and cytokinin sensitivity, crucial for callus pluripotency and organ regeneration. Single-cell RNA sequencing identifies key cell layers and gene interactions in Arabidopsis callus, highlighting the middle cell layer's role in shoot and root regeneration through WOX5/7 and PLT1/2 interactions.20
- Inflorescence-to-floret transition- This study profiles 37,571 rice inflorescence cells using single-cell RNA sequencing, identifying key regulators like DWARF, TILLER1 and OsAUX1. It provides insights into axillary meristem differentiation and floret development, crucial for understanding rice yield determinants. 21
- Nuclei isolation from plant samples : This study demonstrates high-efficiency nucleus isolation and purification methods for ten plant species, verified by flow cytometry. It introduces a precise absolute nucleus counting method using Trucount beads and flow cytometry, and confirms the high quality of nuclear RNA and DNA for advanced molecular research, including single nucleus RNA-seq. 22

- Li S, Simoni Y, Becht E, Loh CY, Li N et al., Human tumor-infiltrating MAIT cells display hallmarks of bacterial antigen recognition in colorectal cancer. Cell Rep Med 2020; 23(1)3:100039 doi: 10.1016/j.xcrm.2020.100039
- Ferreira R, Dopico XC, Oliveira JJ, Rainbow DB, Yang JH et al., Chronic immune activation in systemic lupus erythematosus and the autoimmune PTPN22 Trp620 risk allele drive the expansion of FOXP3+ regulatory T cells and PD-1 expression” found that SLE patients have more CD4+FOXP3+ cells due to thymically-derived Tregs expansion linked to PD-1, IL-2, and soluble SIGLEC-11. Frontiers in Immunol 2019; 10:2606 https://doi.org/10.3389/fimmu.2019.02606
- Alivernini S, MacDonald L, Elmesmari A, Finlay S, Tolusso B, et al. Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis. Nat Med 2020; 8:1295-1306. doi: 10.1038/s41591-020-0939-8. Epub 2020 Jun 29
- Wang Y, Guo L, Yin X, McCarthy EC, et al. Pathogenic TNF-α drives peripheral nerve inflammation in an Aire-deficient model of autoimmunity. Proc Natl Acad Sci USA 2022; 119(4) e2114406119. doi: 10.1073/pnas.2114406119.
- Okamura T, Hamaguchi M, Tominaga H, Kitagawa N, Hoshimoto Y, et al. Characterization of peripheral blood TCR in patients with Type 1 Diabetes Mellitus by BD Rhapsody™ VDJ CDR3 assay. Cells 2022; 11(10):1623. https://doi.org/10.3390/cells11101623
- Rao Y, Zhong D, Qiu K, Cheng D, Li L, et al. Single-cell transcriptome profiling identifies phagocytosis-related dual-feature cells in a model of acute otitis media in rats. Front. Immunol. 2021; 12. https://doi.org/10.3389/fimmu.2021.760954
- May L, Chu C and Zielinkski CE. Single-Cell RNA Sequencing Reveals HIF1A as a Severity-Sensitive Immunological Scar in Circulating Monocytes of Convalescent Comorbidity-Free COVID-19 Patients. Cells; 2024; 12(4):300. https://doi.org/10.3390/cells13040300
- Li R, Li X, Zhao J, Meng F, Yao C, et al. Mitochondrial STAT3 exacerbates LPS-induced sepsis by driving CPT1a-mediated fatty acid oxidation. Theranostics; 2022; 12(2):976-998. doi: 10.7150/thno.63751
- Kwok AJ, Allcock A, Ferreira RC, Canco-Gamez E, Smee M, et al. Neutrophils and emergency granulopoiesis drive immune suppression and an extreme response endotype during sepsis. Nat Immunol. 2023; 24: 767-779 https://doi.org/10.1038/s41590-023-01490-5
- Ma K, Schonnesen A, He C, Cis AY, Sun E, et al. High-throughput and high-dimensional single-cell analysis of antigen-specific CD8+ T cells. Nat Immunol.; 2021; 22; 1590-1598 https://doi.org/10.1038/s41590-021-01073-2
- Mair F, Erickson JR, Frutoso M, Konecny AJ, Greene E, et al. Extricating human tumour immune alterations from tissue inflammation. Nature; 2022; 605(7911):728-735. https://doi.org/10.1038/s41586-022-04718-w
- Yang L, He Y-T, Dong S, Wei XX-U, Chen Z-H, et al. Single-cell transcriptome analysis revealed a suppressive tumor immune microenvironment in EGFR mutant lung adenocarcinoma. J Immunother Cancer; 2022; 10(2):e003534.
- Kim S, Cho E, Kim Y, Han C, Choi BK, et al. Adoptive immunotherapy with transient anti-CD4 treatment enhances anti-tumor response by increasing IL-18Rαhi CD8+ T cells. Nature Comm; 2022; 5314 https://doi.org/10.1038/s41467-021-25559-7
- Wang L, Liu Y, Dai Y, Tang X, Tong Y, et al. Single-cell RNA-seq analysis reveals BHLHE40-driven pro-tumour neutrophils with hyperactivated glycolysis in pancreatic tumour microenvironment. Gut; 2023; 72(5):958-971.
- Lawrence A, Canzi A, Bridlance C, Olivie N, Lansonneur C, et al. Microglia maintain structural integrity during fetal brain morphogenesis. Cell; 2024; 187(4):962-980.e19. https://doi.org/10.1016/j.cell.2024.01.012
- Yu J, Xiao K, Chen X, Hu J, Fu Z, et al. Neuron-derived neuropeptide Y fine-tunes the splenic immune responses. Neuron; 2022; 110(8):P1327-1339. https://doi.org/10.1016/j.neuron.2022.01.010
- Jensen_cody S, Flippo KH, Claflin KE, Yavuz Y, Sapouckey SA, et al. FGF21 Signals to Glutamatergic Neurons in the Ventromedial Hypothalamus to Suppress Carbohydrate Intake. Cell Metab.; 2020; 32(2):273-286. E6. Doi: 10.1016/j.cmet.2020.06.008
- Karahan H, Smith DC, Kim B, Dabin LC, Manun Al-Amin Md, et al. Deletion of Abi3 gene locus exacerbates neuropathological features of Alzheimer’s disease in a mouse model of Aβ amyloidosis. Sci Adv. ; 2021; 7(45):eabe3954. doi: 10.1126/sciadv.abe3954
- Yang, M. C., Wu, Z. C., Chen, R. Y., Abbas, F., Hu, G. B., Huang, X. M., Guan, W. S., Xu, Y. S., & Wang, H. C. (2023). Single-nucleus RNA sequencing and mRNA hybridization indicate key bud events and LcFT1 and LcTFL1-2 mRNA transportability during floral transition in litchi. Journal of Experimental Botany, 74(12), 3613-3629. https://doi.org/10.1093/jxb/erad103
- Zhai, N., & Xu, L. (2021). Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nature Plants, 7(1453–1460). https://doi.org/10.1038/s41477-021-01015-8
- Zong, J., Wang, L., Zhu, L., Bian, L., Zhang, B., Chen, X., Huang, G., Zhang, X., Fan, J., Cao, L., Coupland, G., Liang, W., Zhang, D., & Yuan, Z. (2022). A rice single cell transcriptomic atlas defines the developmental trajectories of rice floret and inflorescence meristems. *New Phytologist*. https://doi.org/10.1111/nph.18008
- Yang, M.-C., Wu, Z.-C., Huang, L.-L., Abbas, F., & Wang, H.-C. (2022). Systematic methods for isolating high purity nuclei from ten important plants for omics interrogation. Cells, 11(23), 3919. https://doi.org/10.3390/cells11233919
Whether you're a seasoned researcher or a newcomer to the field, embarking on a single-cell multiomics experiment requires careful planning, meticulous execution and a keen appreciation for the intricacies of multiomic analysis.
Research Question and Experimental Design
The first step is to clearly define your research question or hypothesis. What specific aspect of cellular biology or disease pathology are you interested in exploring with single-cell multiomics? Are you aiming to identify novel cell types, characterize cellular states or elucidate molecular mechanisms? How many samples/cells do you need to profile to arrive at your desired outcome? To what sequencing depth should you sequence each cell? For example, to characterize subpopulations within a sample you need fewer cells; however, to detect rare cells or do lineage tracing, you need more cells. By articulating your research objectives upfront, you can tailor your experimental design and data analysis strategies to address your scientific goals effectively.
The Satija lab website provides a tool to estimate how many cells you need to sequence so that you see at least “n” number of cells of each type.1
Select Your Experimental Platform
Single-cell multiomics experiments typically involve the integration of multiple omics modalities, such as genomics, transcriptomics, epigenomics, proteomics and metabolomics. You may also have a choice of different technologies to choose from, each having its own pros and cons. Depending on your research question and available resources, you'll need to select the appropriate experimental platforms and technologies for each omics layer. Consider factors such as sensitivity, throughput, cost, hands on time and compatibility with downstream analysis tools when choosing your experimental platform.
The BD Rhapsody™ HT Single-Cell Analysis System offers a microwell-based single-cell partitioning system coupled with a visual workflow QC system.
Sample Preparation and Preservation
Depending on your experimental design, you may need to isolate single cells from tissues, cell cultures or clinical research samples. You may choose manual or automated cell isolation techniques. You may need to enrich or clean up your sample using fluorescence-activated cell sorting (FACS), microfluidic cell sorting, or simple wash and filter. You may also need to preserve your samples using techniques such as cryopreservation, 4 °C preservation and fixation. Careful attention to sample preparation protocols and quality control measures is essential to ensure the reproducibility and accuracy of your results.
Learn more about sample preparation for single-cell multiomics.
Data Analysis and Integration
The analysis of single-cell multiomics data presents unique computational challenges due to the complexity and dimensionality of the datasets involved. Depending on your expertise and resources, you may choose to perform bioinformatics analysis in-house or collaborate with a commercially available computational platform. Key steps in data analysis include quality control, normalization, dimensionality reduction, clustering and integration of omics datasets to identify biological insights and patterns.
Check out the BD Rhapsody™ Sequence Analysis Pipeline tools and find useful resources on our data analysis page.
Interpretation and Validation
Once you've analyzed your single-cell multiomics data, it's time to interpret your findings in the context of your research question and existing literature. What biological insights have you gained from your analysis? Do your results support or challenge existing hypotheses? It's essential to critically evaluate your findings and consider alternative explanations before drawing conclusions. Additionally, experimental validation using orthogonal techniques, such as spatial multiomics, qPCR, FACS or functional assays, can help validate key findings and strengthen the robustness of your results.
Learn more about how to process, normalize, interpret, and validate your single-cell multiomics data on our data analysis blog.
- How many cells do we need to sample so that we see at least n cells of each type? Satija Lab website; accessed July 19, 2024.
For Research Use Only. Not for use in diagnostic or therapeutic procedures.