-
Reagents
- Flow Cytometry Reagents
-
Western Blotting and Molecular Reagents
- Immunoassay Reagents
-
Single-Cell Multiomics Reagents
- BD® OMICS-Guard Sample Preservation Buffer
- BD® AbSeq Assay
- BD® Single-Cell Multiplexing Kit
- BD Rhapsody™ ATAC-Seq Assays
- BD Rhapsody™ Whole Transcriptome Analysis (WTA) Amplification Kit
- BD Rhapsody™ TCR/BCR Next Multiomic Assays
- BD Rhapsody™ Targeted mRNA Kits
- BD Rhapsody™ Accessory Kits
- BD® OMICS-One Protein Panels
- BD OMICS-One™ WTA Next Assay
-
Functional Assays
-
Microscopy and Imaging Reagents
-
Cell Preparation and Separation Reagents
Old Browser
This page has been recently translated and is available in French now.
Looks like you're visiting us from {countryName}.
Would you like to stay on the current location site or be switched to your location?

BD Rhapsody™ Sequence Analysis Pipeline
The BD Rhapsody™ Sequence Analysis Pipeline is a versatile tool that offers the flexibility to run your bioinformatics analysis on either a Seven Bridges cloud-based platform or on a local installation.
The BD Rhapsody™ Sequence Analysis Pipeline:
- Provides a primary analysis of single-cell multiomics data by leveraging cutting-edge algorithms to deliver fast results and deep insights.
- Utilizes an intuitive user interface via a cloud-based platform and is easy to use, regardless of the computational expertise of the user.
- Offers the ability to choose between cloud-based or local installation options and affords maximum convenience and accessibility for single-cell multiomics data analysis.
- Provides broad compatibility of output data with downstream analysis tools such as Seurat and Scanpy.
The .Cellismo output files from the BD Rhapsody™ Sequence Analysis Pipeline can be imported into the BD Cellismo™ Data Visualization Tool for secondary analysis and visualization.
Pipeline Overview
After sequencing, the pipeline takes input from FASTQ files, a reference (Targeted panel or WTA / WTA+ATAC-Seq reference archive), an AbSeq reference (if required) and a supplemental reference (if required) to generate output files and metrics about the pipeline run.
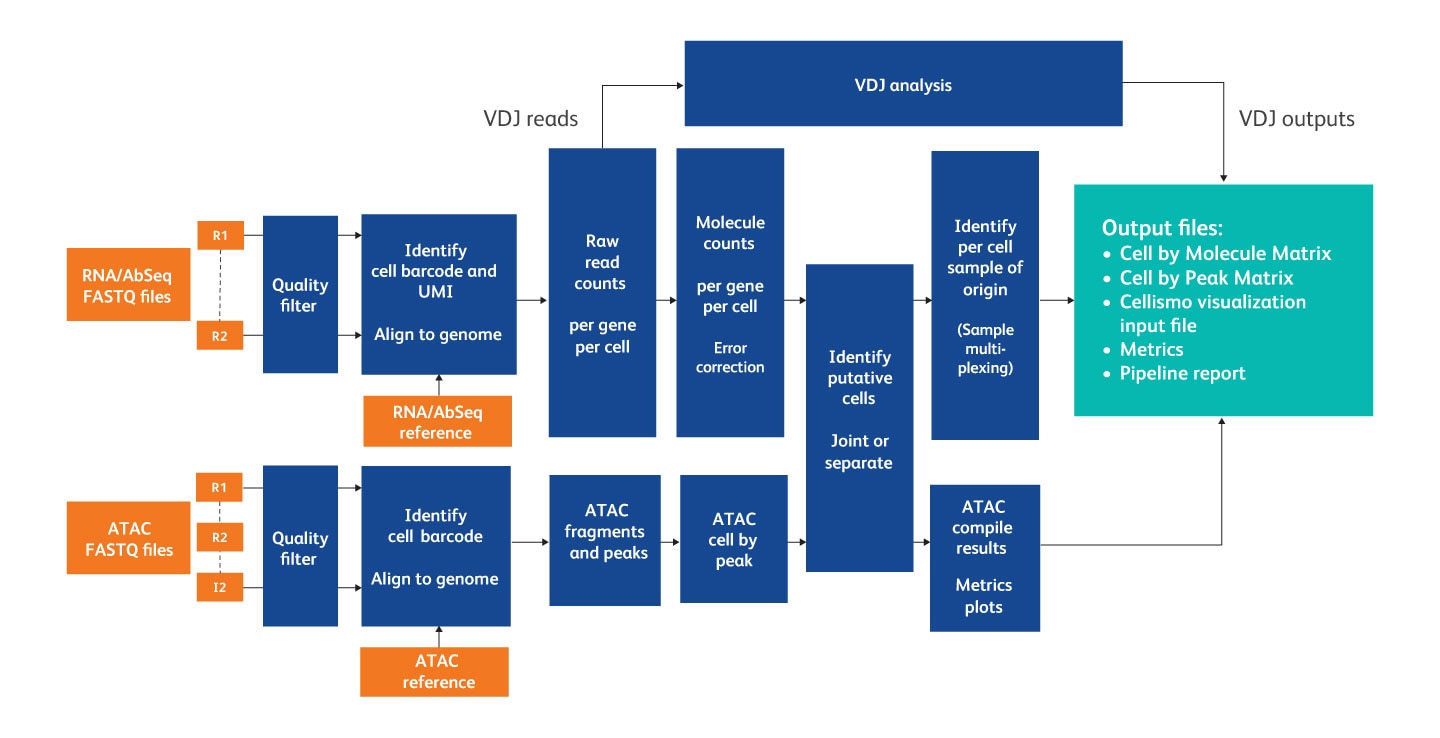
Overview of the steps in the analysis pipeline.
Features
- Free : Upload raw data, run the pipeline and download results from the cloud for free
- Fast: Less than 3 hours to process 1 billion reads
- Simple: One consolidated pipeline for BD Rhapsody™ Whole Transcriptome Analysis Amplification Kit, BD Rhapsody™ Targeted mRNA Kits, BD Rhapsody™ TCR/BCR Next Multiomic Assays and BD Rhapsody™ ATAC-Seq Assays
Release Notes
v3 BD Rhapsody™ Sequence Analysis Pipeline | October 2025
Added
- ATAC:
- Gene Activity output—new modality in the .Cellismo output file and also a separate MEX output file. Gene activity is a Gene-by-Cell matrix, where counts are number of transposase cut sites in the gene body or 2,000 bases upstream of the gene start position
- Transcription factor motif output—new modality in the .Cellismo output file and also a separate MEX output file. This is a TFmotif-by-cell matrix, where values are z-scores of the enrichment of each TF motif
- VDJ
- New assembly algorithm improves speed of this step by up to 23 fold (range 7x-23x), enabling the processing of billions of TCR/BCR reads. Metrics are generally equivalent or slightly better.
- VDJ only pipeline—able to provide only TCR and/or BCR FASTQs and get a cell call and VDJ results. Sample multiplexing with VDJ only is also supported. VDJ in combination with a mRNA assay is still recommended for better cell calling and identification.
- New pipeline node to downsample data to calculate a sequencing saturation curve and median genes per cell curve, which are output on the pipeline report
- Make Rhapsody Reference tool:
- Added an optional input for Transcription Factor Motif PFM file
- Will now filter out readthrough transcripts and genes with only readthrough transcripts. Added optional parameter to turn off this filtering
- Added optional parameter to filter out Y chromosome Pseudo-Autosomal Regions from Human reference build 38
- Pipeline Report:
- New Read Flow diagram, showing a sankey diagram of read filtering steps for each library and for each of the RNA and/or ATAC modalities
- New Sequencing Saturation calculator to enable calculation of required total reads to achieve a target saturation value
Updated
- VDJ
- _VDJ_perCell.csv file CDR3 columns are updated to use CDR3 junction instead of CDR3 alone, resulting in the inclusion of canonical amino acids
- _VDJ_perCell.csv file added full length pairing columns
- New column in AIRR outputs "junction_anchored_aa"—a direct translation of only the CDR3 nucleotide sequence, not influenced by upstream frameshifts
- Update constant region gene identification to prevent mismatched chain types
- Removed PyIR wrapper and call IgBlast directly
- Basic putative cell calling algorithm updated to fix several edge cases and get more precise cell calls. Increase in putative cell number of ~1% is typical. Use of the Expected Cell Count parameter is highly encouraged
- Pipeline Report:
- Various metric alert updates
- Mean bioproducts per cell added to summary section
- Gene expression _MolsPerCell MEX output now contains Ensembl IDs as well as Gene symbols
- Improved library name determination from FASTQ file names
- More aggressive cleanup of polyA sequence in reads to prevent spurious alignments
- Make Rhapsody Reference tool: Extra Sequence input is now included in the BWA-Mem index
- Seven Bridges CWL: Instance types updated to be more performant, and increase size of instances for ATAC related nodes
- ATAC peak annotation now uses transcript features rather than gene features, which better classifies peaks when a gene has multiple transcription start sites
- .Cellismo output file now contains GTF data for genes
- Dimensionality reduction threshold updates: Below 100,000 cells, both t-SNE and UMAP coordinates are generated. Between 100,000 and 300,000 cells, only UMAP coordinates. Above 300,000 cells, a sub-sample of 300,000 cells will be selected and UMAP coordinates generated.
Fixed
- AlignmentAnalysis node was not getting an early cell count estimate, which could cause downstream node scaling issues
- TCR/BCR node failure when the number of valid TCR or BCR reads exceeded 2,147,483,647 reads
- Pipeline Report error when exact cell count parameter specified
- Pipeline Report error when CITE-seq/AbSeq only datasets are run
- Targeted RNA pipeline did not output a DBEC MEX file
- ATAC pipeline could get stuck in QualCLAlign_ATAC for some reference genomes with large numbers of contigs
- Rare issue where an ATAC peak could exceed the length of the contig on which it resides
- Improved handling chromosome names with unexpected characters
- Failure in GenerateSeurat node when there is only 1 AbSeq input
- Rare failure cause by poor quality read 1 data creating a race condition
- Rare failure in ATAC node caused by incorrect BWA-MEM2 binary selection
- ATAC pipeline failure when more than one ATAC library was present in the pipeline inputs
- ATAC pipeline failure when using sample tags or an "Extra seqs" input
- ATAC pipeline discrepancy in putative cell numbers in different output files.
Get Free Access to the Pipeline
Cloud-Based Version
- Go to Velsera.com
- Click Request Access. In the request access window, enter your email address to receive an email invitation to the Seven Bridges Genomics platform within 24 hours.
- Click the link in the email invitation and complete the registration. Seven Bridges Genomics displays the dashboard with the demo projects.
Local Version
- Go to bitbucket.org/CRSwDev/cwl. If necessary, create a Bitbucket account.
- In the left pane, click Downloads > Download Repository. The CWL and YML files will download.
- Unzip the archive. Each folder within the archive is named after the pipeline version to which it corresponds.
-
User's Guide
For Research Use Only. Not for use in diagnostic or therapeutic procedures.