-
Reagents
- Flow Cytometry Reagents
-
Western Blotting and Molecular Reagents
- Immunoassay Reagents
-
Single-Cell Multiomics Reagents
- BD® OMICS-Guard Sample Preservation Buffer
- BD® AbSeq Assay
- BD® OMICS-One Immune Profiler Protein Panel
- BD® Single-Cell Multiplexing Kit
- BD Rhapsody™ ATAC-Seq Assays
- BD Rhapsody™ Whole Transcriptome Analysis (WTA) Amplification Kit
- BD Rhapsody™ TCR/BCR Next Multiomic Assays
- BD Rhapsody™ Targeted mRNA Kits
- BD Rhapsody™ Accessory Kits
- BD® OMICS-One Protein Panels
-
Functional Assays
-
Microscopy and Imaging Reagents
-
Cell Preparation and Separation Reagents
Old Browser
Looks like you're visiting us from {countryName}.
Would you like to stay on the current location site or be switched to your location?
Uncover 30 Human Immune Markers in a Single Experiment
The BD® OMICS-One Immune Profiler Protein Panel (BD® OMICS-One IP Protein Panel) is developed using our antibody-oligo based technology and consists of 30 different specificities against most major human immune markers lyophilized in a single tube.
Designed and optimized to work on the BD Rhapsody™ Single-Cell Analysis System, the BD® OMICS-One IP Protein Panel is tested to work seamlessly with BD Rhapsody™ Single-Cell RNA Assays and BD® Single-Cell Multiplexing Kits to enable CITE-seq studies.
Find more information in the BD® OMICS-One Immune Profiler Protein Panel brochure.

List of BD® OMICS-One Immune Profiler Protein Panel Specificities
30 pre-titrated antibodies
| Specificity | Clone |
|---|---|
| CD3 | UCHT1 |
| CD4 | SK3 |
| CD8 | SK1 |
| CD11c | B-Ly6 |
| CD14 | MΦP9 |
| CD16 | 3G8 |
| CD19 | SJ25C1 |
| CD25 | 2A3 |
| CD27 | M-T271 |
| CD28 | L293 |
| Specificity | Clone |
|---|---|
| CD45RA | HI100 |
| CD56 | NCAM16 |
| CD62L | DREG-56 |
| CD127 | HIL-7R-M21 |
| CD134 | ACT35 |
| CD137 | 4B4-1 |
| CD161 | HP-3G10 |
| CD183 (CXCR3) | 1C6/CXCR3 |
| CD185 (CXCR5) | RF8B2 |
| CD186 (CXCR6) | 13B 1E5 |
| Specificity | Clone |
|---|---|
| CD196 (CCR6) | 11A9 |
| CD197 (CCR7) | 2-L1-A |
| CD272 | J168-540 |
| CD278 | DX29 |
| CD279 | EH12.1 |
| CD357 (GITR) | V27-580 |
| CD366 (TIM-3) | 7D3 |
| HLA-DR | G46-6 |
| IgD | IA6-2 |
| IgM | G20-127 |
Performance
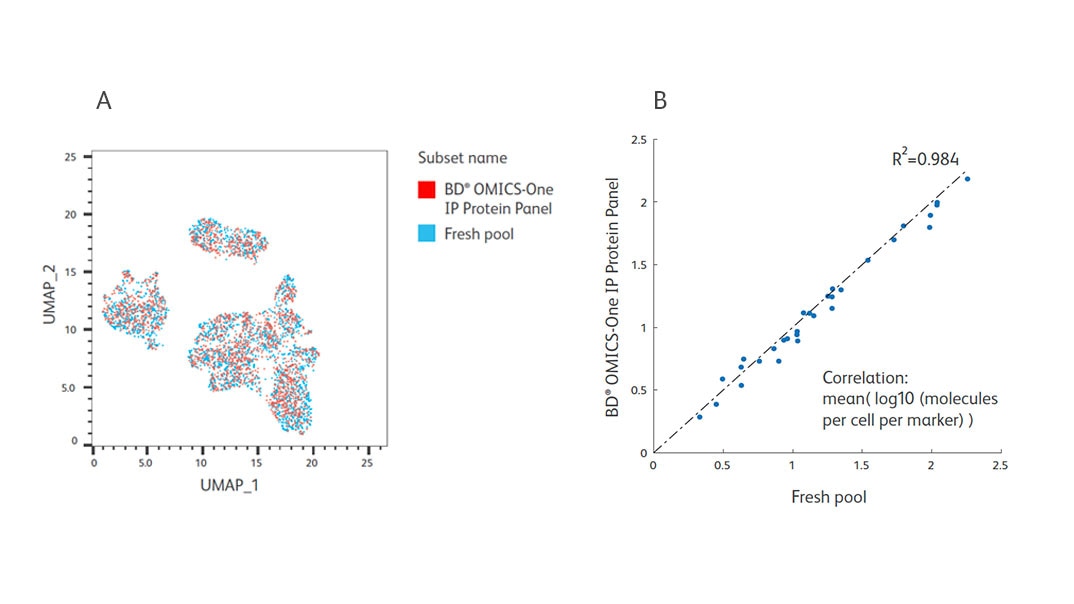
Similar performance between the BD® OMICS-One IP Protein Panel versus freshly mixed BD® AbSeq Antibodies. Following isolation from whole blood, peripheral blood mononuclear cells (PBMC) were split into resting (untreated) and activated (treated with CD3/CD28 for 24 hours) groups. A 1:1 mixture of the resting and activated cells were stained with either the BD® OMICS-One IP Protein Panel or a freshly prepared mixture of the same AbSeq specificities. Equal number of cells were then loaded onto the BD Rhapsody™ Cartridges and AbSeq and WTA libraries were generated and sequenced (n = 2 individual experiments for this study). Data were analyzed using SeqGeq™ Software.
A. UMAP demonstrated strong overlap in the cell groups identified between the BD® OMICS-One IP Protein Panel and fresh BD® AbSeq Antibody-stained samples.
B. The total number of AbSeq molecules detected by the BD® OMICS-One IP Protein Panel and fresh BD® AbSeq Antibody mixture showed a high correlation with R2 greater than 0.98.
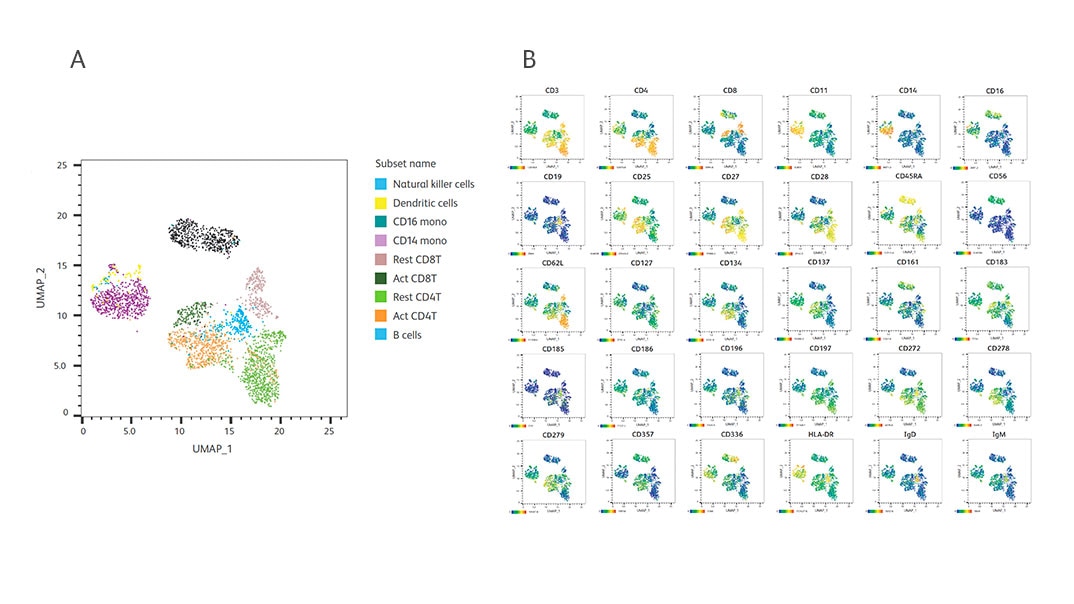
Performance of all 30 AbSeq specificities included in the BD® OMICS-One IP Protein Panel. PBMCs were activated and prepared as described in the figure above. After staining, cells were captured on the BD Rhapsody™ System and AbSeq and WTA libraries were generated and sequenced. To obtain over 80% sequencing saturation, the libraries were sequenced at 20,000 reads/cell for WTA and 30,000 reads/cell for AbSeq using the Illumina™ NextSeq™ High-Output Kit. Data were analyzed using SeqGeq™ Software. We repeated the above experiments with at least two different donors. The representative figures from one donor are shown here.
A. Cell annotation of UMAP of resting + activated PBMCs resolved by the BD® OMICS-One IP Protein Panel antibodies and the WTA mRNA profile.
B. Heat maps of each AbSeq clone from BD® OMICS-One IP Protein Panel on UMAP from (A) showing the specificity of AbSeq detection for individual cell type.
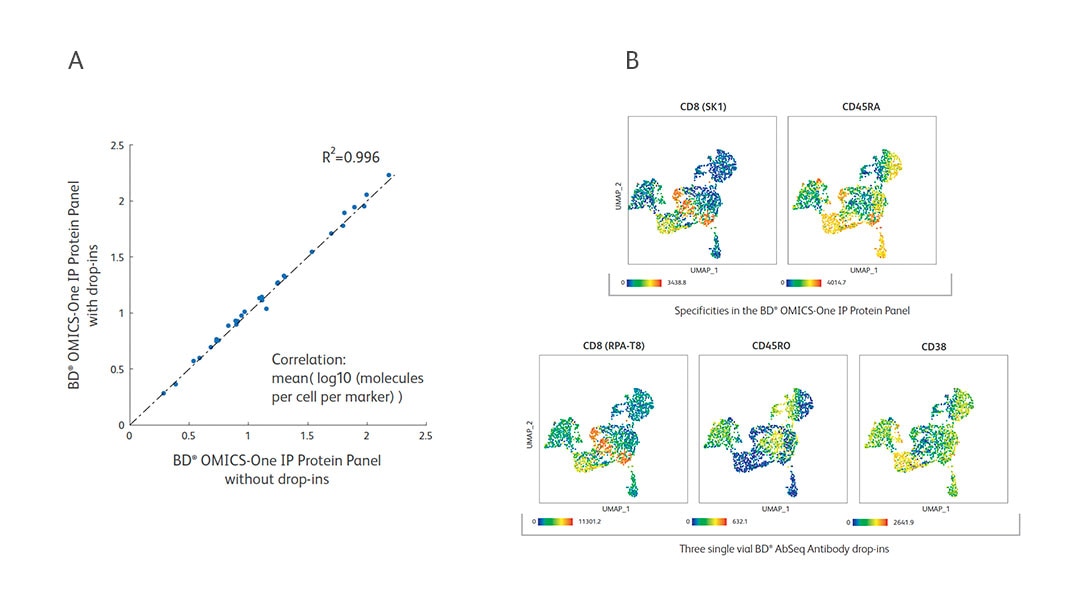
The BD® OMICS-One IP Protein Panel is a flexible backbone panel and accommodates additional AbSeq specificities. Three BD® AbSeq Antibodies were added and mixed with the reconstituted BD® OMICS-One IP Protein Panel pellet (n = 2).
A. The BD® OMICS-One IP Protein Panel performance was not impacted by drop-ins as shown by high correlation (R2 over 0.99 with or without drop-ins).
B. The BD® OMICS-One IP Protein Panel specificities of CD8 (SK1) and CD45RA (top row) were assessed against the specificity of drop-ins CD8 [RPA-T8], CD45RO and CD38 (bottom row) and are shown in UMAP. Drop-in for CD38 detected cell types that are expected to be positive for CD38. Drop-in clone for CD8 (RPA-T8) showed a staining pattern very similar to the BD® OMICS-One IP Protein Panel clone (SK1) suggesting the high specificity of drop-in antibody as well as the compatibility of two clones for the same antigen. The contrasting expression pattern of CD45RO (drop-in) compared to CD45RA (BD® OMICS-One IP Protein Panel) further confirmed that adding the AbSeq specificities to the BD® OMICS-One IP Protein Panel had no adverse impact on experimental outcomes.
The BD® OMICS-One IP Protein Panel is designed to work with RNA and multiplexing assays
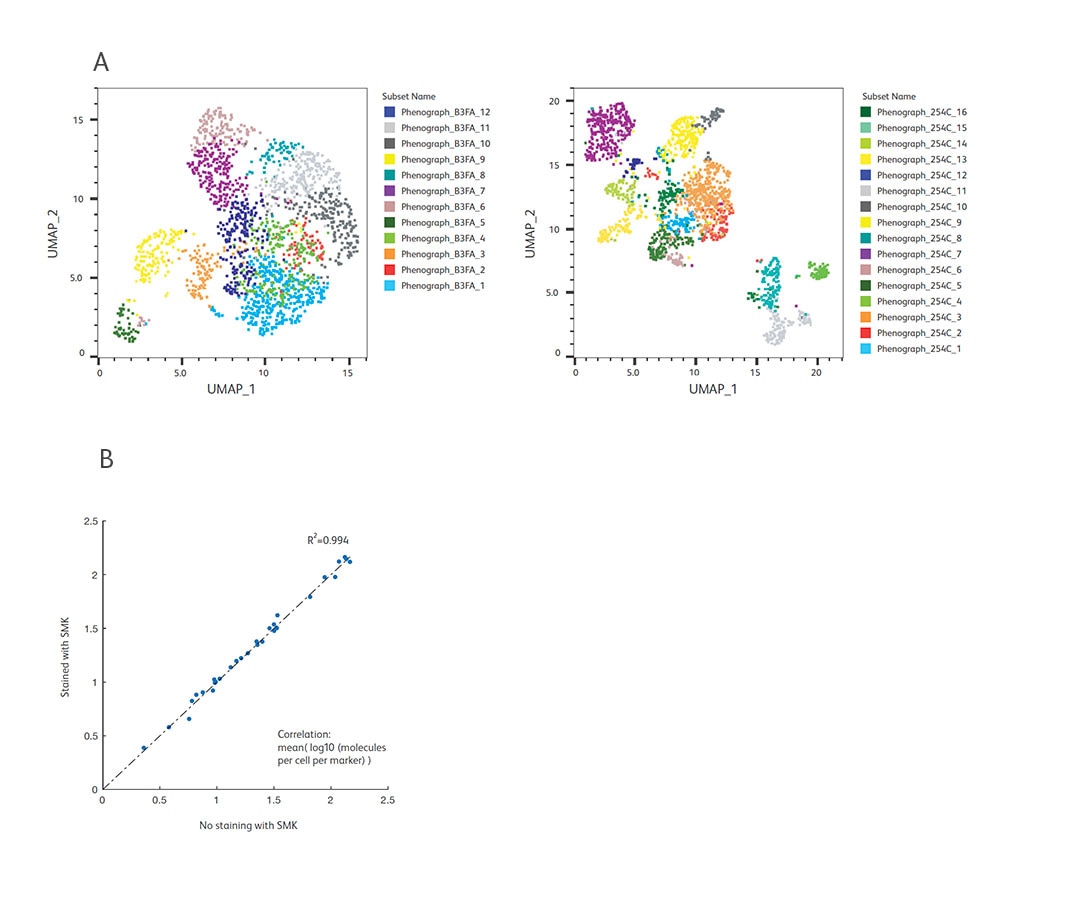
A. WTA and AbSeq libraries from BD® OMICS-One IP Protein Panel-stained cells (1:1 mixture of resting and activated PBMCs) were generated and sequenced. To illustrate the power of multiomic analysis, we analyzed the WTA data only (mRNA analysis) and compared with WTA + AbSeq data (mRNA and protein analysis). UMAP coordinates and unbiased clustering (Phenograph) using only WTA (mRNA) data are shown on the left, while coordinates and annotations using WTA + AbSeq (mRNA and protein) data are shown on the right. With a multiomics approach, additional cell types were revealed offering deeper biological insights.
B. To test the compatibility of the BD® Single-Cell Multiplexing Kit (SMK) and the BD® OMICS-One IP Protein Panel, we performed cell staining with the SMK and the BD® OMICS-One IP Protein Panel together and generated WTA, AbSeq and SMK libraries for sequencing. The expression of markers in the BD® OMICS-One IP Protein Panel was then compared to data generated in the absence of the SMK. These data showed that addition of the SMK does not impact the BD® OMICS-One IP Protein Panel as demonstrated by high correlation (R2 >0.99) between the BD® OMICS-One IP Protein Panel + WTA versus BD® OMICS-One IP Protein Panel + WTA + SMK.
For Research Use Only. Not for use in diagnostic or therapeutic procedures.