-
Training
- Flow Cytometry Basic Training
-
Product-Based Training
- FACSAria Product Based Training
- FACSMelody Product-Based Training
- FACSLyric Product-Based Training
- FACSCanto Product-Based Training
- LSRFortessa Product-Based Training
- FACSymphony Product-Based Training
- FACSDuet Product-Based Training
- HTS Product-Based Training
- BD FACSDiscover™ S8 Cell Sorter Product Training
-
Advanced Training
-
Thought Leadership
-
Product News
- Blogs
- Scientific Publications
-
Events
- Nature Research Academies Workshop 2023
- CYTO 2023: Advancing the World of Cytometry
- Singapore Gene & Cell Therapy Conference 2023
- EuroFlow Educational Workshop
- Nature Research Masterclass 2023
- Novel Approaches to Single-Cell Plant Research: from Real-Time Imaging Cell Sorting to Single-Nuclei Transcriptomics
- Advances in Immune Monitoring Series
-
Product News
-
- FACSAria Product Based Training
- FACSMelody Product-Based Training
- FACSLyric Product-Based Training
- FACSCanto Product-Based Training
- LSRFortessa Product-Based Training
- FACSymphony Product-Based Training
- FACSDuet Product-Based Training
- HTS Product-Based Training
- BD FACSDiscover™ S8 Cell Sorter Product Training
-
- Nature Research Academies Workshop 2023
- CYTO 2023: Advancing the World of Cytometry
- Singapore Gene & Cell Therapy Conference 2023
- EuroFlow Educational Workshop
- Nature Research Masterclass 2023
- Novel Approaches to Single-Cell Plant Research: from Real-Time Imaging Cell Sorting to Single-Nuclei Transcriptomics
- Advances in Immune Monitoring Series
- Singapore (English)
-
Change country/language
Old Browser
This page has been recently translated and is available in French now.
Looks like you're visiting us from {countryName}.
Would you like to stay on the current country site or be switched to your country?
BD Pharmingen™ Recombinant Human Active Caspase-3
(RUO)





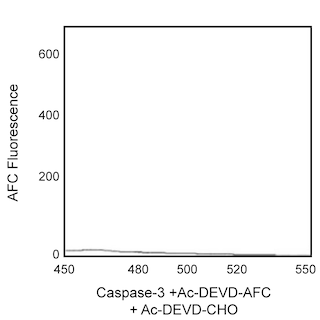
Rate of Ac-DEVD-AFC hydrolysis using 2.5 ng/ml recombinant human active caspase-3 (CPP32). The rate of enymatic hydrolysis was measured at an emission of 505 nm upon excitation at 400 nm using a Perkin-Elmer LS50B fluorometer equipped with a thermostated plate reader.

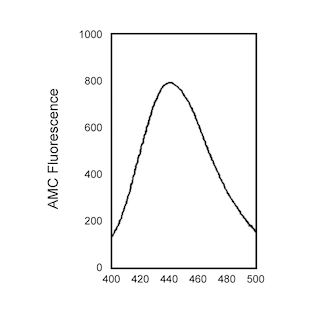
Cleavage of the Ac-DEVD-AMC substrate by recombinant human caspases-3, -6, -7, and -8. The activity of the caspases was analyzed by spectrofluorometry using an excitation at 380 nm and an emission wavelength of 430-460 nm (peak is at 440 nm). The concentration of each caspase (50, 100, or 1000 ng/ml) used in the reactions is noted in the graphs.


BD Pharmingen™ Recombinant Human Active Caspase-3

BD Pharmingen™ Recombinant Human Active Caspase-3

Recombinant Human Active Caspase-3
Regulatory Status Legend
Any use of products other than the permitted use without the express written authorization of Becton, Dickinson and Company is strictly prohibited.
Description
Caspases are cysteine proteases that play a central role in apoptosis. The caspase family was discovered by searching human cDNA libraries for sequences homologous to ced-3, a C. elegans death gene that is required for normal apoptosis during development. The first mammalian homolog of ced-3 to be identified was ICE (interleukin-1β-converting enzyme). Subsequent numerous human ced-3 homologues were rapidly identified which led to multiple names for many of the molecules. To achieve consistency, "caspase" was adopted as a root name for all family members. The name was selected based on two catalytic properties of these enzymes, the "c" reflects a cysteine protease mechanism and "'aspase" refers to their unique ability to cleave after aspartic acid. There are at least 10 members, caspase-1 (ICE), caspase-2 (ICH-1), caspase-3 (CPP32, Yama, apopain), caspase-4 (TX, ICH-2, ICErel-II), caspase-5 (ICErel-III), caspase-6 (Mch2), caspase-7 (Mch3, ICE-LAP3, CMH-1), caspase-8 (MACH, FLICE, Mch5), caspase-9 (ICE-LAP6, Mch6), and caspase-10 (Mch4). Each caspase is synthesized as an inactive proenzyme that is processed by cleavage at asparte residues by another protease or by self-proteolysis. The processed forms consist of large (17-22 kDa) and small (10-12 kDa) subunits which associate to form an active enzyme. The activation of some of these caspases has been shown to occur during apoptosis.
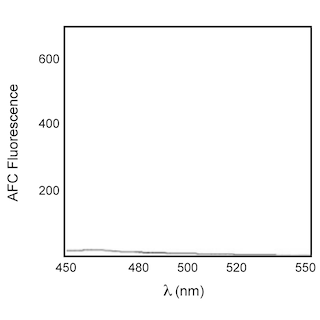
Caspase-3, -6, -7, and -8 have been shown to play a role in apoptosis induced by the death receptors Fas and tumor necrosis factor receptor type 1 (TNFR1). One of their substrates is poly (ADP ribose) polymerase (PARP). PARP is an enzyme that is involved in DNA repair and genomic maintenance. Activated caspases 3, 6, 7 and 8 can all cleave PARP from its 116 kDa form to an 85 kDa residual fragment. The cleavage separates the DNA-binding domain in the N-terminus of PARP from its C-terminus catalytic domain, and results in loss of normal PARP function. The cleavage site in PARP is C-terminal to Asp-216. The upstream sequence of the PARP cleavage site, DEVD (Asp-Glu-Val-Asp), is utilized as a basis for highly specific caspase-3 substrates such as Ac(N-acetyl)-DEVD-AFC (7-amino-4-trifluoromethylcourmarin) and Ac(N-acetyl)-DEVD-AMC (7-amino-4-methylcoumarin) as well as the caspase-3 aldehyde inhibitor Ac-DEVD-CHO.
Preparation And Storage
The thawed active enzyme is generally stable at 4°C for about one week.Active caspase-3 was expressed in E.coli and purified. When expressed in E. coli, caspase-3 will spontaneously undergo autoprocessing to yield the subunits characteristic of the active enzyme.
Recommended Assay Procedures
Enzyme Evaluation: The rate of caspase enzymatic hydrolysis is measured by release of AFC, from the Ac-DEVD-AFC caspase substrate, through emission at 480-520 nm (peak at 505 nm) upon excitation at 400 nm using UV spectrofluorometry. The rate of enzymatic hydrolysis can also be measured by release of AMC, from the Ac-DEVD-AMC caspase substrate, through emission at 440 nm upon excitation at 380 nm.
This protocol is used to measure caspase-3 enzymatic activity using the synthetic fluorogenic peptide, Ac-DEVD-AMC, as the caspase enzyme substrate. The enzyme cleaves the substrate between D and AMC, releasing the fluorescent AMC. AMC release is measured by spectrofluorometry using UV excitation wavelength of 380 nm and emission wavelength of 440 nm.
1. Add 20 µM of Ac-DEVD-AMC to 1 ml of assay buffer (20 mM PIPES, 100 mM NaCl, 10 mM DTT, 1 mM EDTA, 0.1% (w/v) CHAPS, 10% sucrose, pH 7.2). Add active caspase-3 to the mixture to a final concentraton of 50 ng/mL.
2. Incubate for 1 hour at 37°C.
3. Measure the AMC liberated from Ac-DEVD-AMC using a spectrofluorometer with an excitation wavelength of 380 nm and an emission wavelength of 440 nm.
Product Notices
- Since applications vary, each investigator should titrate the reagent to obtain optimal results.
- Please refer to www.bdbiosciences.com/us/s/resources for technical protocols.
Companion Products
Please refer to Support Documents for Quality Certificates
Global - Refer to manufacturer's instructions for use and related User Manuals and Technical data sheets before using this products as described
Comparisons, where applicable, are made against older BD Technology, manual methods or are general performance claims. Comparisons are not made against non-BD technologies, unless otherwise noted.
For Research Use Only. Not for use in diagnostic or therapeutic procedures.
Report a Site Issue
This form is intended to help us improve our website experience. For other support, please visit our Contact Us page.