-
Training
- Flow Cytometry Basic Training
-
Product-Based Training
- BD FACSDiscover™ S8 Cell Sorter Product Training
- Accuri C6 Plus Product-Based Training
- FACSAria Product Based Training
- FACSCanto Product-Based Training
- FACSLyric Product-Based Training
- FACSMelody Product-Based Training
- FACSymphony Product-Based Training
- HTS Product-Based Training
- LSRFortessa Product-Based Training
- Advanced Training
-
- BD FACSDiscover™ S8 Cell Sorter Product Training
- Accuri C6 Plus Product-Based Training
- FACSAria Product Based Training
- FACSCanto Product-Based Training
- FACSLyric Product-Based Training
- FACSMelody Product-Based Training
- FACSymphony Product-Based Training
- HTS Product-Based Training
- LSRFortessa Product-Based Training
- United States (English)
-
Change country/language
Old Browser
This page has been recently translated and is available in French now.
Looks like you're visiting us from {countryName}.
Would you like to stay on the current country site or be switched to your country?



Introducing BD FACSuite™ Application Version 1.6
Next-generation application featuring BD ElastiGate™ Algorithm- machine learning enabled auto-gating algorithm
Preserve Your Precious Samples for up to 72h at 4°C
BD® OMICS-Guard Sample Preservation Buffer—a one step, flexible buffer optimized to preserve your samples, compatible with flow cytometry, RNA-seq, CITE-seq and qPCR

Automate More.
Get More Done.
Drive efficiency from sample to answer with the power of the all-new BD FACSDuet™ Premium Sample Preparation System

New Blue-Laser Fluorochromes are Here
Improve data resolution with BD Horizon RealBlue™ 705 and BD Horizon RealBlue™ 744 Reagents

Visualize the Future of Flow Cytometry
Derive greater insights and unlock new discoveries using image-enabled spectral sorting.


Applications and Solutions
A comprehensive suite of trusted products, integrated solutions, useful tools and a wealth of information to advance your flow cytometry applications.
Innovative solutions to power your diagnostics and research
Backed by cutting-edge technology and more than 45 years of flow cytometry expertise
“My order was placed quickly and easily. No technology problems. The new BD Biosciences website worked flawlessly! It saved me a lot of time. The new experience is like taking a walk in the park. It’s like night and day!”

“My order was placed quickly and easily. No technology problems. The new BD Biosciences website worked flawlessly! It saved me a lot of time. The new experience is like taking a walk in the park. It’s like night and day!”
“I’m really excited about the BD Horizon RealBlue™ 780 Dye. The staining we have done so far with Ki-67 and Granzyme B can be seen very clearly, even with high dilutions.”

“I’m really excited about the BD Horizon RealBlue™ 780 Dye. The staining we have done so far with Ki-67 and Granzyme B can be seen very clearly, even with high dilutions.”
“BD is positioned with BD® CS&T and BD® FC Bead technology to enable instrument standardization simply from day to day, instrument to instrument, and lab to lab. Reference control–based instrument standardization is the next step for flow cytometry.”

“BD is positioned with BD® CS&T and BD® FC Bead technology to enable instrument standardization simply from day to day, instrument to instrument, and lab to lab. Reference control–based instrument standardization is the next step for flow cytometry.”
“At last, a fully integrated end-to-end walkaway solution, which is going to make our lives significantly easier. It reduces errors in both the pre-analytical and analytical phases, leading to increased productivity and efficiency, but most importantly provides a comprehensive fully audit-able account of the whole process. Finally, a solution that creates capacity to allow us to meet our key performance indicators.”

“At last, a fully integrated end-to-end walkaway solution, which is going to make our lives significantly easier. It reduces errors in both the pre-analytical and analytical phases, leading to increased productivity and efficiency, but most importantly provides a comprehensive fully audit-able account of the whole process. Finally, a solution that creates capacity to allow us to meet our key performance indicators.”
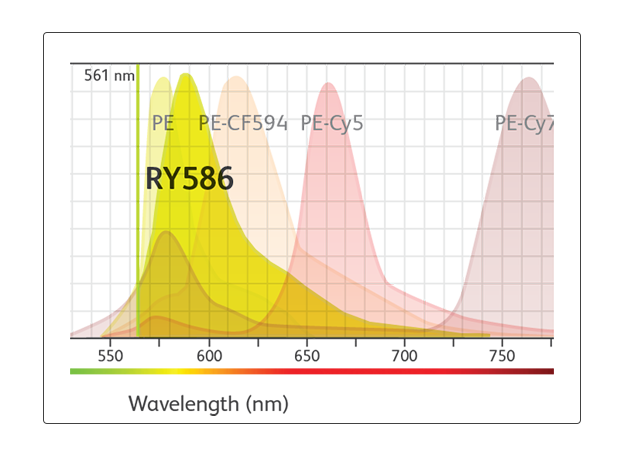
"BD Horizon RealYellow™ 586 (RY586) Reagents look like the best alternative for PE and will simplify panel design on conventional flow cytometers. They can also be an additional dye for spectral panel design.”

"BD Horizon RealYellow™ 586 (RY586) Reagents look like the best alternative for PE and will simplify panel design on conventional flow cytometers. They can also be an additional dye for spectral panel design.”


Utilize our featured resources with invaluable information to advance your science.
Explore a variety of tools for your multicolor flow cytometry, from panel design to selection tools—an array of aids to support your research.
Be part of the flow cytometry community with the latest flow cytometry news, thought leader opinions, blogs on breakthrough research, interesting flow cytometry publication reviews, and more.
Use our excellent product and technical support with our >45 years of flow cytometry expertise and support resources.
Find a seamless customer experience and an easy way to create and manage accounts.
Explore a variety of ways that work with your purchasing process to place and track orders.
Explore our webinar series spanning a broad range of topics and in-depth reviews by senior scientists, professionals and clinicians.
Use a variety of training courses for instruments and applications to take full advantage of the capabilities of BD products.
Report a Site Issue
This form is intended to help us improve our website experience. For other support, please visit our Contact Us page.